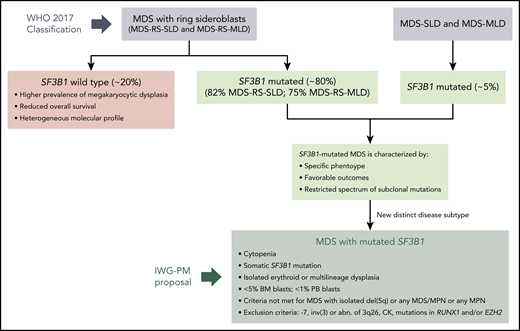
In this issue of Blood, Malcovati et al (for the International Working Group for the Prognosis of Myelodysplastic Syndromes [IWG-PM]) propose a new subtype of myelodysplastic syndrome (MDS) characterized by somatic mutations in SF3B1.1
Definition of a distinct MDS disease subtype characterized by somatic mutations in SF3B1. MDS patients with somatic SF3B1 mutations share a common phenotype, with favorable outcomes and a restricted spectrum of subclonal mutations, independently of their WHO 2017 category. Malcovati et al propose a new MDS subtype characterized by SF3B1 mutations, following the classification criteria depicted in the figure. Abn, abnormalities; BM, bone marrow; CK, complex karyotype (≥3 chromosomal alterations); MLD, multilineage dysplasia; MPN, myeloproliferative neoplasms; PB, peripheral blood; SLD, single-lineage dysplasia.
Definition of a distinct MDS disease subtype characterized by somatic mutations in SF3B1. MDS patients with somatic SF3B1 mutations share a common phenotype, with favorable outcomes and a restricted spectrum of subclonal mutations, independently of their WHO 2017 category. Malcovati et al propose a new MDS subtype characterized by SF3B1 mutations, following the classification criteria depicted in the figure. Abn, abnormalities; BM, bone marrow; CK, complex karyotype (≥3 chromosomal alterations); MLD, multilineage dysplasia; MPN, myeloproliferative neoplasms; PB, peripheral blood; SLD, single-lineage dysplasia.
MDSs are a group of clonal hematopoietic stem cell disorders characterized by ineffective hematopoiesis, myelodysplasia, peripheral cytopenias, and potential for clonal evolution. Recurrent gene mutations are present in ≥90% of MDS cases.2 These mutations affect pathophysiological features of MDS and play a role in their clinical heterogeneity. Mutations also contribute to more precise classification and risk stratification of the patients.3 However, the genetic basis of MDS is complex. The spectrum of mutations is heterogeneous, the driver genes are not specific for MDS, and some of these mutations can be present in individuals with clonal hematopoiesis of indeterminate potential (CHIP).4 Therefore, the diagnosis of MDS remains heavily reliant on morphology. Although there is no doubt that morphology will continue to represent a fundamental step in the diagnostic process, closer integrartion of morphology and molecular profiles are needed in many hematological malignancies.
In the revised 2017 World Health Organization (WHO) Classification, MDS with isolated del(5q) remains the only MDS subtype defined by a genetic abnormality, although SF3B1 mutation has been included as a diagnostic criterion in MDS with ring sideroblasts (MDS-RS).5 More specifically, a diagnosis of MDS-RS (with single- or multilineage dysplasia) can be made if RS comprise ≥15% of nucleated erythroid cells or if ≥5% RS plus an SF3B1 mutation are present. Mutations in the splicing factor SF3B1 occur in 25% of all MDS cases but affect >80% of MDS-RS.6,7 In addition, they are independent predictors of favorable outcomes in MDS.8 This is a clear example of a genotype-phenotype relationship and supports the inclusion of SF3B1 mutation as a diagnostic criterion in MDS-RS. Patients with MDS-RS are characterized by ineffective erythropoiesis, erythroid dysplasia, low blast percentage, and generally favorable outcomes. However, the inclusion of SF3B1 mutations in the WHO classification raised new questions: Is the phenotype and outcome of these patients really associated with the presence of RS or with the presence of SF3B1 mutations? How have the 15% and 5% cutoffs been defined? Do these cutoffs discriminate patients with unique features? For example, in patients with RS, SF3B1 mutations identify 2 groups of patients with clearly different outcomes, but the presence of >15% RS does not have an impact on SF3B1-mutated MDS.9
In this report, Malcovati et al, writing for the IWG-PM, provide evidence supporting the recognition of SF3B1-mutant MDS as a distinct diagnostic entity and further validate this proposal by interrogating the IWG-PM data set.1 This data set includes 3479 patients with known SF3B1 mutation status and represents the largest MDS data set with genetic data reported to date. Thus, prior reports of the correlation of SF3B1 mutations with clinical phenotype in MDS are strengthened by the validation in this large data set.
As reviewed by the authors, several lines of evidence support recognition of somatic SF3B1 mutations, and not the presence of RS, as a disease-defining feature (see figure). First, SF3B1 mutations often represent a founding genetic lesion, with several lines of evidence consistent with this mutation being a driver event. Second, SF3B1 is a major determinant of disease phenotype. Specifically, (1) SF3B1-mutant MDS has ineffective erythropoiesis with erythroid dysplasia and RS; (2) in SF3B1-mutant MDS with multilineage dysplasia, very mild dysplasia in granulocytic or megakaryocytic lineages is present, which lacks clinical significance; (3) the presence of excess of blasts significantly affects survival; (4) there is a female prevalence; and (5) there is no difference in survival in SF3B1-mutant MDS depending on the RS percentage or the number of dysplastic lineages. Third, SF3B1 mutations have an independent prognostic value on survival and risk of progression to acute myeloid leukemia. SF3B1 mutations predict a favorable outcome in very low and low Revised International Prognostic Scoring System categories and in sideroblastic categories, but they have no impact in MDS with excess blasts. In addition, the authors explore the prognostic value of co-occurring cytogenetic abnormalities and somatic mutations in SF3B1-mutant MDS. Fourth, SF3B1 mutations may predict response to specific agents, such as luspatercept.
Considering all the above, Malcovati et al propose the following classification criteria for MDS with mutated SF3B1 (see figure): (1) cytopenia defined by standard hematologic values; (2) somatic SF3B1 mutation; (3) isolated erythroid or multilineage dysplasia; (4) bone marrow blasts <5% and peripheral blood blasts <1%; and (5) WHO criteria for MDS with isolated del(5q), myelodysplastic/myeloproliferative neoplasm with RS and thrombocytosis or other myelodysplastic/myeloproliferative neoplasms, and primary myelofibrosis or other myeloproliferative neoplasms are not met. The following genetic lesions represent exclusion criteria: monosomy 7, inv(3) or abnormalities of chromosome 3q26, complex karyotype (≥3 chromosomal alterations), and co-occurring mutations in RUNX1 and/or EZH2.
The presence of an SF3B1 mutation on its own is not sufficient to diagnose MDS, since SF3B1 mutations have been reported in other hematological and nonhematological cancers7 and in individuals with CHIP.4 In patients with unexplained cytopenia, SF3B1 mutations are highly predictive of developing MDS-RS,10 although prospective studies are needed to validate these observations and establish the predictive value of SF3B1-mutated clones in this context.
Patients with RS but no SF3B1 mutation constitute a heterogeneous group of patients with less favorable outcomes. If SF3B1-mutant MDS is recognized as a distinct entity, additional studies will be needed to define how to best reclassify these patients. The presence of specific genetic lesions might be useful to assess their prognosis.
Our understanding of hematological malignancies is inevitably associated with the progress of molecular genetics. Therefore, the inclusion of molecular data has the potential to significantly improve diagnostic and prognostic assessment. The SF3B1 paradigm is a clear example of that, as captured in this Special Report by Malcovati et al. However, although the evidence supports the distinction of SF3B1-mutant MDS as a disease subtype, morphology remains essential to classify these patients. Thus, an integrated diagnosis, including genetic data as well as morphological, biological, and clinical parameters, is necessary.
Conflict-of-interest disclosure: The authors declare no competing financial interests.