Decision making in the public healthcare system is heavily invested in screening and preventative medicine in addition to the translation of national initiatives that help in implementing precision medicine. Molecular testing for cancers and inherited disorders helps families, patients and healthcare providers in disease management. Different genome programs and genetics initiatives around the world are contributing and providing data to help decision makers, pharmaceutical companies and other health related commercial entities to tailor diagnosis and treatment in a healthcare era that honors molecular individuality and population differences.
The Saudi Human Genome Program (SHGP) is a national program committed to sequencing 100,000 genomes. In its early stages, it focused on understanding monogenic disorders. The data output was then expanded to allow researchers to interrogate the genetics of complex disorders and understand the molecular landscape of this highly inbred population. The results of the first phase of this program (consisting 35000 WES and gene panels and 2000 WGS) was translated into the establishment of a knowledge database of population specific mutations and single nucleotide variants (SNVs) that is used for molecular diagnosis. This was translated to a definitive molecular diagnosis in nearly half of ~2500 cases with Mendelian monogenic disorders using 13 targeted gene panels. To date; the SHGP knowledge database contains more than 2000 disease causing mutations and more that 4 million SNVs.
In the Kingdom, there are two established national screening programs, premarital and newborn. These programs are extremely powerful at serving patients and carriers/family members equally, and will further benefit from the availability of population specific molecular databases. As we soar towards the mid-phase of the program, it has become obviously a valuable platform to inform clinical decision and policy making. The presumed presentation of carriers and family members in light of autosomal recessive inheritance is indicative of possible missed diagnosis when they are asymptomatic or present with milder clinical features, creating heavier burdens on the healthcare system. As further evidence emerges, genomics will continue to change practice in other healthcare areas including cancer and chronic diseases.
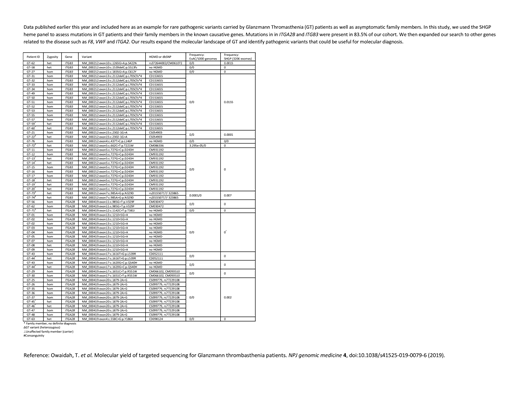
In this study, we present rare genetic variants in common and rare bleeding disorders, where we have looked at data from the SHGP with both disease stratification and blindly to redefine and classify the rarity of pathogenic genetic variants in associated genes. We assessed an initial set of 1400 individuals using a targeted gene panel consisting of 393 genes implicated in nonmalignant blood related disorders (SHGP heme panel). We looked for variants and mutations to reclassify bleeding disorders based on the incidence of carriers and affected individuals. We then used a replication set of ~5000 WES to validate pathogenicity and assess allele frequencies. Our data will help impact decision making for effective screening and prevention of common blood related disorders and will aid in increasing the diagnostic yield, identifying predisposition markers and implementing better genetic counseling programs.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal