Drug resistance is a major cause of concern in cancer chemotherapy that not only reduces efficacy but also causes toxicity through overdosing. Multiple myeloma (MM) is the second-most common haematological malignancy in the USA that remains a challenging disease to cure due to the presence of significant complexity and heterogeneity at molecular level with high treatment cost. Proteasome inhibitor, the standard of care drug, has proven to remarkably improve myeloma patient outcomes but is associated with dose limiting toxicities and drug resistance. Therefore, new combination treatment regimens are urgently needed for the treatment of PI-resistant MM that can lower the required dose of standard-of-care drug without compromising on treatment efficacy.
We have created an optimization-regularization based computational prediction method called secDrug that uses large-scale pharmacogenomics databases to identify novel secondary drugs against PI-resistance when used as single agent or in combination. We applied secDrug to PIs-resistant cell lines (>100) in the Genomics of Drug Sensitivity in Cancer (GDSC) database and predicted the top drugs that can be best combined with PIs to overcome PI-resistance in MM. These include: Survivin inhibitor (YM155), Nicotinamide phospho ribosyl transferase or Nampt inhibitor (FK866), PIKfyve inhibitor (YM201636), Raf inhibitor (PLX-4720), Bcl2 inhibitor (Navitoclax),HSP90 inhibitor (17-AAG), AKT inhibitor (KIN00102), HDAC inhibitors (Panobinostat, SAHA), S6K1-specific inhibitor (PF-4708671), and the neddylation inhibitor (MLN4924).
To validate our prediction results, we treated human multiple myeloma cell lines (HMCLs) highly resistant to the proteasome inhibitors Bortezomib, Carfilzomob, Oprozomib and Ixazomib and >10 clonally derived acquired resistant cells with the top predicted best secondary drugs. Next, we treated three pairs of parental PI-sensitive (P) and clonally-derived PI-resistant (R) cell lines from PI-sensitive (P) HMCLs representing acquired PI-response with the top predicted drug combinations. Our preliminary in vitro data confirmed our in silico predictions of secondary drugs based on secDrug analysis.
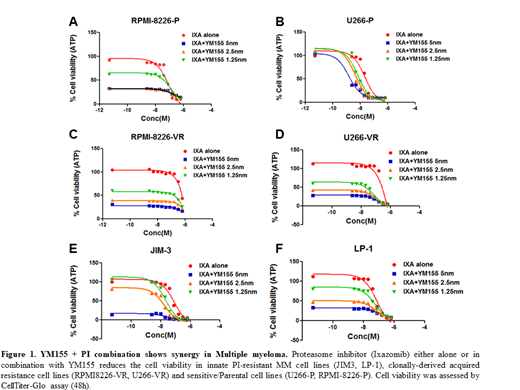
Among these, YM155 and YM155 + Ixazomib showed strikingly very high in vitro cytotoxicity against PI- resistant MM cell lines (Figure 1). The results, based on Chou-Talalay's combination index (CI) theorem, show that this combination works highly synergistically where the IC50 of ixazomib in MM cells was significantly reduced in presence of YM155. Similar results were obtained for the other top predicted combination regimens. Our findings provide a strong case for combining YM155 with ixazomib to enhance sensitivity or overcome resistance to ixazomib and thereby improve patient outcome. Further, this study provides additional support towards using secDrug for predicting secondary drugs in cancers resistant to the standard-of-care therapy. Currently, we are exploring the molecular mechanisms behind the synergism between proteasome inhibitors and the top drugs identified by our algorithm as candidates for secondary combination therapies using next-gen transcriptomics and single-cell analysis approaches. Earlier studies have shown that YM155 potentially induces selective cell death in human pluripotent stem cells. Further, the presence of cancer stem cells sub population/ MM-CSCs in drug-resistant tumors have been shown to significantly contribute to the development of PI-resistance in MM. Hence, we propose that YM155 reverses PI-resistance by reducing the stem cell load in multiple myeloma cell line model system. Therefore, our future plan is to assess the effects of YM155 on MM-cancer stem cells like subclones/ sub population/ MM-CSCs including CD19- CD138- quiescent stem cells, ALDH+ and side populations/SP) in drug-resistant tumors.
Overall, our in vitro validation results corroborated with our in silico prediction of secondary drugs.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal