Background
Diffuse large B-cell lymphoma (DLBCL) is a genetically and clinically heterogeneous disease. The cell-of-origin (COO) classification subdivides DLBCL into the transcriptionally defined activated B-cell (ABC) and germinal center B-cell (GCB) subtypes. Recently, 2 novel classifiers based on genetic features were independently proposed further unraveling the diversity of DLBCL [Schmitz et al, NEJM2018; Chapuy et al, Nat Med2018]. The concordance between the 2 novel classification systems has not yet been systematically studied. However, both classifiers are largely complementary to COO subtypes, and describe overlapping genotypes.
We previously demonstrated the feasibility of COO classification by noninvasive plasma genotyping in a limited gene panel using Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) [Scherer et al, STM2016], and this approach has now been replicated by others. In this study, we take first steps toward a comprehensive non-invasive classification of novel DLBCL genetic subtypes using a limited gene panel.
Methods
We analyzed genetic profiling of 476 DLBCL patients reported by Schmitz et al (NEJM 2018) as a training set to build 2 classifiers in a limited gene panel applicable to plasma genotyping from CAPP-Seq: (1) A COO classifier (i.e. ABC, GCB and Unclassified); (2) A comprehensive genetic classifier (i.e. EZB, BN2, MCD, N1 and Other as defined in Schmitz et al, NEJM 2018). Features were limited to genetic alterations captured by our plasma genotyping panel, and those with population frequency of at least 10% in at least one genetic subtype. Our final model comprised 100 features: 64 recurrently mutated genes, 26 amplifications, 7 deletions and 3 translocations (BCL2, BCL6 and MYC). After cross-validation in the training set, we applied the 2 classifiers to the dataset from Chapuy et al (Nat Med 2018) as well as pretreatment plasma genotyping data from patients previously reported by our group [Kurtz et al, JCO 2018].
Results
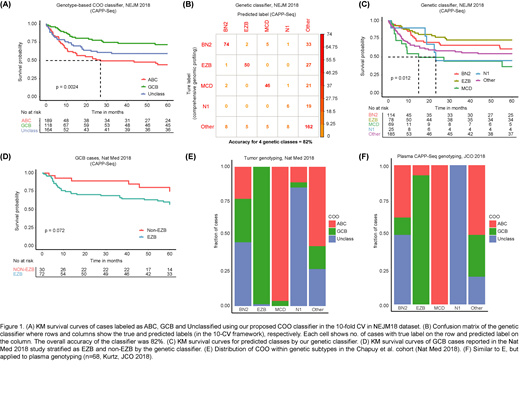
We first evaluated our 2 classifiers in a 10-fold cross-validation (CV) framework in the NEJM 2018 dataset of Schmitz et al. Despite modest performance of our GCB/ABC classification, COO labels had the expected significant prognostic associations (Fig. 1A). Overall accuracy of our second classifier to determine novel genetic subtypes was 82% (Fig. 1B). Consistent with the original study, inferred MCD and N1 subtypes had adverse prognosis compared to EZB and BN2 (Fig. 1C).
We next applied our classifiers to the Chapuy et al (Nat Med 2018) dataset. Again, consistent with findings by Schmitz et al (NEJM 2018), the EZB subset of GCB cases had inferior outcome compared to non-EZB cases (Fig. 1D). We next examined the cross-correlation between the two classifiers and observed the expected enrichment patterns of ABCs in the MCD subset and enrichment of GCBs in the EZB subset (Fig. 1E).
Finally, we applied our classifiers to plasma genotyping data previously reported by our group [Kurtz et al., JCO 2018]. We restricted the analysis to cases with a mean variant allele fraction ≥0.5% (n=68). Similar to the original study, 59% of cases (40/68) were labeled unclassifiable (i.e. Other). We compared the distribution of COO subtypes within the Schmitz genetic clusters. Representation of ABC and GCB within the clusters inferred from Plasma genotyping (Fig. 1F) was similar to the distribution from Tumor genotyping (Fig. 1E).
Conclusions
We describe 2 new classifiers applicable to noninvasive plasma genotyping data that recapitulate transcriptionally and genetically defined DLBCL subtypes. Using independent datasets, we show the feasibility of classification with a limited feature set with good prediction accuracy and prognostic stratification of defined subtypes. Genotyping of pretreatment plasma samples suggest that comprehensive non-invasive classification of genetic subtypes of DLBCL is achievable.
Kurtz:Roche: Consultancy. Diehn:BioNTech: Consultancy; Quanticell: Consultancy; Roche: Consultancy; AstraZeneca: Consultancy; Novartis: Consultancy. Alizadeh:Roche: Consultancy; Genentech: Consultancy; Janssen: Consultancy; Pharmacyclics: Consultancy; Gilead: Consultancy; Celgene: Consultancy; Chugai: Consultancy; Pfizer: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal