Introduction: Azacitidine (AZA) is a DNA hypomethylating agent used in myeloid neoplasms, however approximately half of patients show treatment failure or relapse. Last years, several studies have showed that genetic mutations may influence on response and survival of the treated patients. Other biomarkers that have traditionally been associated with the response to AZA are the recovery of the platelet count and the presence of abnormalities in the chromosome 7. Finally, the methylation dynamics of genes promoters could be a useful tool to predict the clinical response.
Aim: To assess the predictive value on response to AZA of clinical features, cytogenetics, genetic mutations and the methylation dynamics of CDKN2B and DLC-1 promoters in a series of patients with myeloid neoplasms.
Methods: We studied 85 patients with myelodysplastic syndromes (MDS) and 28 patients with acute myeloid leukemia (AML) who received AZA. Treatment schedules included 5-azacytidine 75 mg/m2/day for 7 days (n=62) and 5-azacytidine 75 mg/m2/day for 5 days (n=51). According to the International Working Group (IWG) 2006 criteria for MDS, responders were defined as patients achieving complete remission (CR), bone marrow CR (mCR), partial remission (PR) or stable disease with hematologic improvement (SDHI). Sixteen genes with prior implication in the pathogenesis of myeloid diseases were analyzed: NPM1, FLT3, CEBPA, TET2, DNMT3A, IDH1, IDH2, ASXL1, EZH2, RUNX1, SETBP1, TP53, SF3B1, SRSF2, U2AF1 and ETNK1. For methylation assay, EZ DNA Methylation-Gold kit (Zymo Research) was employed for the bisulfite treatment of DNA. Methylation of CDKN2B and DLC-1 were qualitatively studied pre- and post-AZA with unmethylated and methylated primers and analysis of the PCR products in a QIAxcel System (Qiagen). Overall survival (OS) was measured from start of AZA treatment until the last follow-up or death from any cause. P values <0.05 were considered as statistically significant.
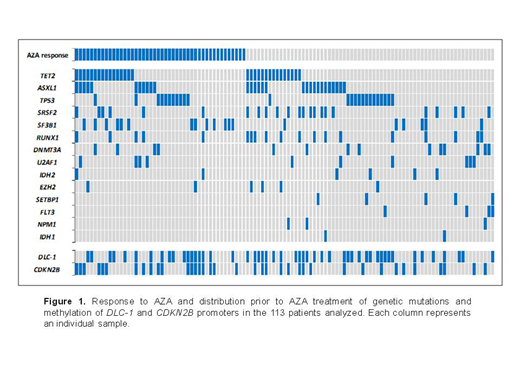
Results: Of the 113 patients analyzed, 46 patients (41%) showed response to AZA (median cycles=6): CR=19, mCR=6, PR=12, and SDHI=9. Stable disease and treatment failure were observed in 20 (17%) and 47 patients (42%), respectively. Analysis of clinical features showed that platelet doubling after the first AZA cycle was significantly associated with a better response (58% vs. 31% responders, P=0.041). Forty-seven patients (42%) had chromosomal abnormalities. A positive co-occurrence between del(7q)/-7 and del(17p) was found (n=6, P<0.001) and was significantly associated with a worse response to AZA (P=0.039). Pre-treatment, genetic mutations were presented in 98 patients (87%) and methylation of CDKN2B and DLC-1 promoters were detected in 50 (44%) and 37 (33%) patients respectively (Figure 1). Patients with SF3B1 mutations showed a better response to AZA (68% vs. 35% responders, P=0.008) and a higher OS (30 vs. 15 months, P=0.019) than the remaining patients. In contrast, patients with mutations in transcription factors (RUNX1, SETBP1, NPM1) showed a worse response to AZA (20% vs. 47% responders, P=0.014) and a lower OS (10 vs. 19 months, P=0.010). Splicing genes mutations (SF3B1, SRSF2 and U2AF1) were related to favourable karyotypes (90% vs. 59%, P<0.001) and a lower DLC-1 methylation (31% vs. 57%, P=0.006). Interestingly, patients with DLC-1 methylation showed higher levels (>5%) of bone marrow blasts (63% vs. 40%, P=0.016), higher frequency of complex karyotypes (40% vs. 15%, P=0.002), a positive co-occurrence with TP53 alterations (38% vs. 16%, P=0.009) and its methylation dynamics showed a strong trend towards predict the response to AZA: 57% of patients without DLC-1 methylation or who reduced it post-treatment responded favourably to the drug and 65% of patients who maintained or increased it did not respond (P=0.053). In addition, leukemia transformation was higher in MDS patients with DLC-1 methylation post-AZA than in the remaining MDS patients (58% vs. 21%, P=0.002).
Conclusion: The combination of chromosomal alterations and genetic mutations together with the monitoring of platelet count and DLC-1 methylation could contribute to give light in myeloid neoplasms to the heterogeneity observed both in the response to AZA and in patient survival.
Samples and data from patients included in this study were provided by the INCLIVA Biobank (PT13/0010/0004).
Hernandez Boluda:Incyte: Other: Travel expenses paid.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal