Leukemic patients with MLL gene rearrangements (MLL-r) suffer from unfavorable prognosis and lack of effective targeted therapy. Recent studies suggest that MLL-r leukemias are frequently associated with epigenetic dysregulations; therefore, targeting epigenetic machineries may provide therapeutic opportunities to improve outcomes for MLL-r leukemias. Tudor domains are chromatin reader modules that recognize the methylated-lysine/arginine on histones. Given the crucial roles of histone methylations in controlling gene expression, we hypothesized that certain Tudor domain-containing genes may play essential roles in MLL-r leukemias.
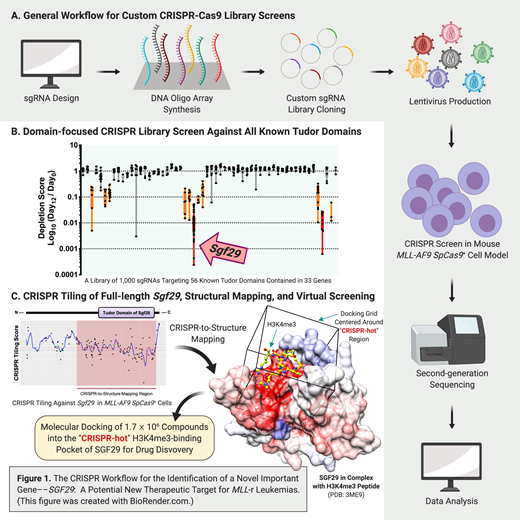
To identify a Tudor domain-containing gene(s) that is crucial for MLL-r leukemic cells, we custom-built a CRISPR library of 1,000 sgRNAs that targets the exon regions of all known Tudor domains (total 56 Tudor domains in 33 genes) with an average density of 17.7 sgRNAs per Tudor domain (Fig. 1A & B). We delivered this sgRNA library by lentiviral transduction into mouse MLL-AF9 leukemic cells constitutively expressing Streptococcus pyogenesCas9 (SpCas9). We then compared the frequencies of each integrated sgRNA sequence before versus after 12 days of culture using high-throughput sequencing (NextSeq, Illumina), and identified Sgf29 (also known as Ccdc101) as a novel essential gene for MLL-r leukemia. Using SpCas9-mediated gene depletion (individual sgRNA co-expressed with a TagRFP reporter) and high-throughput flow cytometric growth competition assay (Attune NxT, ThermoFisher), we validated that SGF29 is essential in both mouse (MLL-AF9) and human (MV4-11 and MOLM-13) MLL-r leukemic cells. We also observed a stronger dependency to SGF29 in MLL-r leukemic cells compared to other cell types including T-cell lymphoma (Hut78), colon cancer (SW620), breast cancer (MDA-MB-231), lung cancer (H661) and glioblastoma (U251) using the growth competition assay. This suggests that targeting SGF29 may represent a novel therapeutic opportunity for MLL-r leukemias.
Mechanistically, we found that CRISPR depletion of Sgf29 in MLL-AF9 cells induced morphological differentiation by Wright-Giemsa staining, and resulted in up-regulation of Cd11b (a myeloid-differentiation marker) and down-regulation of c-Kit (a hematopoietic progenitor and leukemic stem cell marker) by flow cytometry. To identify functionally important regions of Sgf29, we performed a saturation CRISPR-Cas9 gene body scan (also known as CRISPR-tiling screen) that targets all available "NGG" protospacer adjacent motifs (PAM) along the Sgf29 exons (total 147 sgRNAs; average targeting density of 6.0 bp per sgRNA) in MLL-AF9SpCas9+cells (Fig. 1A & C). After high-throughput sequencing of the integrated sgRNA sequences, we mapped the data onto a three-dimensional (3D) crystal structure of SGF29 (PDB: 3ME9) and found that the CRISPR-scan hotpots (i.e.the essential regions) are centered around the H3K4me3-recognition pocket of SGF29. Using RNA-seq and gene set enrichment analysis (GSEA), we observed that the proto-oncogene Myc and Myc-regulated gene sets were significantly down-regulated in the Sgf29 depleted condition. Importantly, ectopic expression of Myc cDNA significantly rescued the lethality induced by Sgf29 depletion. Our current analyses suggest that SGF29 may serve as an upstream regulator of MYC to support the continuous proliferation of the leukemic cells.
Based on our 3D CRISPR-scan analysis, we reasoned that development of small molecules binding to the "CRISPR-hot" region may disrupt the normal function of SGF29 (Fig. 1C). Therefore, we performed a virtual screen by docking 1.7 million diverse compounds into this H3K4me3-recognizing pocket of SGF29 using AutoDock Vina (The Scripps Research Institute). By combining the CRISPR-tiling screen, 3D structural analysis, and in silico compound docking, we prioritized a list of candidate binders of SGF29 Tudor domain for in vitro binding and cell survival assays. These may pave the way to develop the first SGF29 inhibitor with the ability to suppress MLL-r leukemias.
Taken all together, our data suggest the epigenetic reader protein, SGF29, is a novel therapeutic candidate for MLL-r leukemias. We anticipate that the Tudor domain-focused CRISPR dropout screening and CRISPR-tiling strategies can also be applied to identify therapeutically relevant Tudor domain-containing genes in other hematopoietic malignancies.
Armstrong:Janssen: Research Funding; OxStem Oncology: Consultancy, Equity Ownership; Accent Therapeutics: Consultancy, Equity Ownership; Syros Pharmaceuticals: Consultancy, Equity Ownership; Imago Biosciences, Inc.: Consultancy, Equity Ownership; Epizyme, Inc.: Consultancy, Equity Ownership; AstraZeneca: Research Funding; Novartis: Research Funding; Mana Therapeutics: Consultancy, Equity Ownership; Cyteir Therapeutics: Consultancy, Equity Ownership; C4 Therapeutics: Consultancy, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal