Background: As recurrence of acute myeloid leukemia (AML) is difficult to predict, it is important to detect it by measuring minimal residual disease (MRD). PML-RARA, RUNX-RUNX1T1, CBFB-MYH11 are regarded as the reliable MRD markers. However, in AML with normal karyotype and many other forms, no MRD markers have been established. NPM1 mutations, occurring in approximately 30% of adult AML cases, and 50-60% of AML cases with normal karyotype, represent one of the most frequent mutations in AML. Recently, NPM1 mutation is reported to be useful in assessing MRD. We undertook a retrospective and prospective investigation of the usefulness of NPM1 mutation as an MRD marker in Japanese patients with AML.
Methods: The subjects were 38 NPM1-mutated AML patients with first hematological remission at several hospitals related to our institution between 2001 and 2018. This study was approved by the ethics committee of Nippon Medical School and the informed consents were obtained from all patients, according to the Declaration of Helsinki. We analyzed peripheral blood cells or bone marrow cells at diagnoses, and evaluated only bone marrow cells after diagnoses. Detection of NPM1 mutation was carried out using allele-specific real time PCR following creation of a complementary primer. After dilution of the samples, sensitivity to TCTG, CATG, and CCTG was found to be 0.001%. The NPM1 mutant copies were qualified only at successful amplification of internal control.
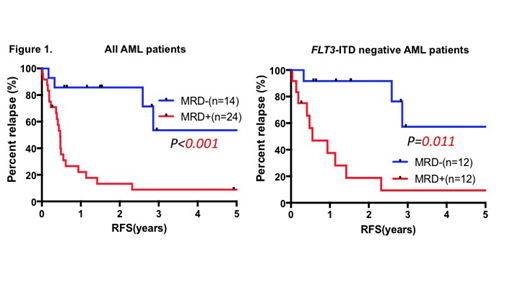
Results: The median age of the patients was 58 years (18-79 years). There were 32 cases with intermediate cytogenetic prognosis and 6 cases with unclear chromosomal profile. Of the 38 cases, 14 cases (37%) were FLT3-ITD-positive and allogeneic hematopoietic stem cell transplantation was carried out in 14 cases (37%). The base sequence was TCTG in 36 cases and CCTG in 2 cases. Persistence of NPM1-mutatation was present in 25 patients with first hematological remission (66%). Compared with patients with MRD negative, patients with MRD positive were associated with DNMT3A mutation (MRD positive 12/25 vs MRD negative 0/13, p=0.003). The rate of relapse in patients with MRD positive was significantly higher than those of in patients with MRD negative (MRD positive 76% vs MRD negative 23%, p=0.004). The rates of relapse free survival (RFS) and overall survival (OS) in patients with MRD positive were significantly lower than those in patients with MRD negative (RFS at 2 years: MRD positive 14% vs MRD negative 86% p=0.003; Figure 1, OS at 2 years: MRD positive 25% vs MRD negative 93%, p<0.001). In FLT3-ITD negative group, the rates of RFS in patients with MRD positive were significantly lower than those in patients with MRD negative. (RFS at 2 years: MRD positive 21% vs MRD negative 92% p=0.001; Figure 1).
Conclusion: The presence of MRD with NPM1 mutation is significantly associated with relapse and it is useful to decide their treatment strategy. Especially, there is the usefulness of NPM1 mutation as an MRD marker in NPM1 positive Flt3-ITD negative AML patients who are generally classified as favorable risk. According to previous reports, it is known that NPM1-mutated AML sometimes relapse with losing NPM1 mutations. However, in this study, all NPM1-mutated AML relapse without losing NPM1 mutations. We need to collect more patients and are going to confirm whether there are patients who relapse with losing NPM1 mutations or not. We plan to analyze the genetic background of MRD positive and negative patients with next-generation sequencing. We are going to announce the genetic characteristics in addition to this result at ASH.
Usuki:Astellas Pharma Inc: Research Funding, Speakers Bureau; Daiichi Sankyo Co., Ltd.: Research Funding, Speakers Bureau. Kako:Bristol-Myers Squibb: Honoraria; Pfizer Japan Inc.: Honoraria. Inokuchi:Bristol-Myers Squibb: Honoraria, Research Funding; Novartis: Honoraria; Celgene: Honoraria; Pfizer: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal