BACKGROUND:
Comprehensive genomic profiling (CGP) performed by next-generation sequencing of DNA detects genomic alterations including point mutations, insertions/deletions, copy number variations, and select gene rearrangements. When RNA sequencing is included in CGP, it allows for expanded detection of gene fusions, which are common in hematologic malignancies and sarcomas. When such tumors involve bone, a decalcification step is frequently employed to soften tissues prior to processing and sectioning. While commonly used acid-based decalcification methods work quickly, the resulting nucleic acid damage can be profound. In this study, we examine the effects of decalcification on DNA and RNA sequencing in the clinical setting.
DESIGN:
1711 consecutive formalin-fixed paraffin embedded samples were evaluated by CGP during routine clinical care via DNA and RNA sequencing, using a hybrid-capture next-generation sequencing assay (FoundationOne®Heme). Specimen site [e.g. bone/ bone marrow or soft tissue] and decalcification status were extracted from pathology reports and H&E review. Samples were considered decalcified if reported as such in the pathology report or if visible decalcified bone was present on the H&E. Samples documented to be processed with fixatives other than formalin were excluded. Sequencing failures were defined as samples that failed DNA extraction (DNAx), RNA extraction (RNAx), or library construction (LC) due to insufficient nucleic acid to advance into sequencing. Samples were only evaluated for RNA if DNAx was successful (1594 cases).
RESULTS:
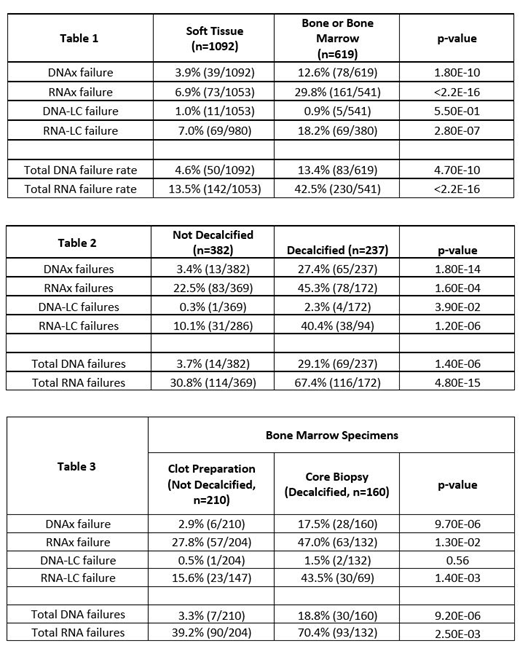
Specimen site was a strong predictor of sequencing failure, with a significant increase in failure rate from bone/bone marrow samples (n=619) compared to samples from soft tissue sites (n=1092) for both DNA (13.4% vs 4.6%, p=4.7E-9) and RNA (42.5% vs 13.5%, p<2.2E-16).
Of the bone/bone marrow samples, 237 of 619 samples were decalcified. Decalcification was associated with significantly higher failure rates than non-decalcified samples for both DNA (29.1% vs 3.7%) and RNA (67.4% vs 30.8%) (Table 2).
One method of avoiding decalcification for bone marrow samples is utilization of clot preparations, where aspirates are processed as an FFPE block. Clot preparations fail sequencing significantly less often than decalcified core biopsies (DNA: 3.3% vs 18.8%, p=9.2E-06; RNA: 39.2% vs 70.4%, p=2.5E-03) (Table 3).
CONCLUSIONS:
CGP of samples acquired from bone and bone marrow sites is challenging, with a lower success rate for DNA and RNA sequencing than soft tissue sites. The higher overall failure rate correlates with use of decalcification agents leading to degradation of nucleic acids and impacts RNA sequencing significantly more than DNA (67.4% vs 30.8% failed). Clot preparations of bone marrow samples performed better than core biopsies for both DNA and RNA. The higher overall RNA sequencing failure rates still observed in in non-decalcified bone/bone marrow are predominantly due to RNA failure of non-decalcified clot preparations. These samples likely have increased failure rates secondary the use of non-standard fixatives (e.g. B+, Bouin's, AZF, etc.) not documented in the pathology report and the frequency of hypocellular clot preparations in conjunction with higher requirements for RNA yield compared to DNA yield.
To increase CGP success rates, decalcification should be avoided when possible. Peripheral blood and bone marrow aspirate samples rarely fail sequencing (<1%, data not shown) and are preferable to decalcified samples if adequate tumor is present. Bone marrow clot preparations perform better than bone marrow core biopsies and clot preparations should be fixed with 10% neutral buffered formalin. If decalcification is required for processing, EDTA based decalcification methods and/or minimizing decalcification times is recommended.
Duncan:Foundation Medicine, Inc.: Employment. Danziger:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Duncan:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Hemmerich:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc.: Employment. Edgerly:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc: Employment. Huang:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc.: Employment. Vergilio:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Elvin:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. He:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Britt:Foundation Medicine, Inc: Employment. Reddy:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc: Employment. Sathyan:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Alexander:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Ross:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc.: Employment. Brown:Foundation Medicine, Inc.: Employment; F. Hoffman La Roche, Ltd.: Equity Ownership. Ramkissoon:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc.: Employment. Severson:F. Hoffman La Roche, Ltd.: Equity Ownership; Foundation Medicine, Inc.: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal