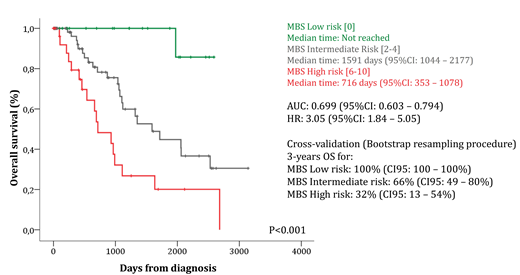
Background: The recent efforts to uncover the molecular heterogeneity of myelodysplastic syndromes (MDS), mainly by new sequencing technologies, allow the comprehensive identification of driver mutations and/or altered gene expression recurrently found in a recognizable fraction of patients. Ongoing efforts are being made to clarify the impact of molecular changes on clinical phenotype and prognosis, as well as their role in the pathogenesis of MDS. Refining risk stratification allows the proposition of risk-adapted therapy and may shed light in biology of MDS. Aims: Based on the gene expression of selected metabolic targets, we aimed to design a score system that improves MDS overall survival prediction. Patients and methods: Clinical, mutations and transcriptomic data from CD34+ cells from 159 MDS patients and 17 healthy volunteers freely available at Gene Expression Omnibus (GEO/NCBI: GSE58831) were used in the present work. Forty-one genes related to metabolic processes, previously demonstrated as deregulated among diverse neoplastic conditions, were ranked and asked for differential expression and prognostic impact. Each gene was dichotomized according to Receiving-Operating Curve (ROC) and Cox Proportional-Hazard Model was used for multivariate analysis using gender, age and IPSS-R as cofounders. Genes independently associated with overall survival (OS) were selected to compose the Molecular-Based Score (MBS) and integer weight of each one was defined according Hazard Ratio (HR). Survival curves were constructed using Kaplan-Meyer method and compared with Log-Rank Test. ROC c-statistic was used to measure the predictive function of MBS. Prediction accuracy of MBS was cross-validated by a nonparametric bootstrap procedure with 1,000 resamplings of the original cohort allowing replacement and also estimated their respective 95% confidence interval (95% CI) computing the bias-corrected and accelerated bootstrap interval. Results: Among selected genes, 18 were differentially expressed between CD34+ cells from MDS and healthy volunteers. Fifteen genes predict OS in univariate analysis, of which ACLY (HR: 0.48; 95%CI: 0.24 - 0.96; P=0.04), ANPEP (HR: 2.16; 95%CI: 1.08 - 4.31; P=0.02), PANK1 (HR: 0.43; 95%CI: 0.19 - 0.98; P=0.04), PKM (HR: 2.01; 95%CI: 1.02 - 3.93; P=0.04) and SLC25A5 (HR: 0.52; 95%CI: 0.27 - 0.99; P=0.05) were independently associated with OS. Higher expression of ANPEP and PKM, as well as lower expression of ACLY, PANK1 and SLC25A5 were considered to integer high risk being attributed weight 2 for each condition. MBS varied from 0 to 10 (median=2) and was calculated as: MBS Low-Risk =0 (MBS-LR; n=28); MBS Intermediate-Risk=2 and 4 (MBS-IR; n=90) and High-Risk: ≥6 (MBS-HR; n=48). The modeled MBS showed a ROC c-statistic of 0.699 (95%CI: 0.603 - 0.794) and HR=3.05 (95%CI: 1.81 - 5.05; P<0.001) and efficiently identified patients with different risk: MBS-LR (3-year OS: 100%; median time [MT]: not reached); MBS-IR (3-year OS: 66% [95%CI: 53% - 83%]; MT: 52.2 months [95%CI: 34.2 - 71.5]) and MBS-HR (3-year OS: 32% [95%CI: 17% - 61%]; MT: 23.5 months [95%CI: 11.6 - 35.4]). Using Cox Proportional Hazard Model for multivariate analysis, the proposed MBS (HR:2.5 [95%CI: 1.2-4.96]; P=0.008) was independently associated with OS using gender (HR:0.48 [95%CI: 0.21-1.06]; P=0.07), age (HR:1.03 [95%CI: 1.001-1.07]; P=0.02) and IPSS-R (HR:0.98 [95%CI: 0.69-1.36]; P=0.88) as confounders. The bootstrap resampling procedure validated the MBS and demonstrated the stability of its prediction (3-year OS for MBS-LR: 100% [95%CI: 100-100%]; MBS-IR: 66% [95%CI: 49-80%]; MBS-HR: 32% [95%CI: 13-54%]; P<0.001). Conclusions: The proposed Molecular-Based Score (MBS) independently predict OS with superior efficacy in comparison to the most clinically relevant prognostic factors, and reinforce the therapeutic opportunity of metabolic processes in MDS.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal