Acute Lymphoblastic Leukemia is the most prevalent hematological neoplasm in Indian children and around 85% are of B-cell origin (B-Acute Lymphoblastic Leukemia (B-ALL)). ETV6-RUNX1 (E/R) is one of the common translocations seen in B-ALL. We have identified Insulin like Growth Factor 2 mRNA Binding Protein 1 (IGF2BP1), an oncofetal protein, to be specifically overexpressed in bone marrow samples of E/R positive B-ALL patients. We have tried to characterize the role of this protein in B-ALL.
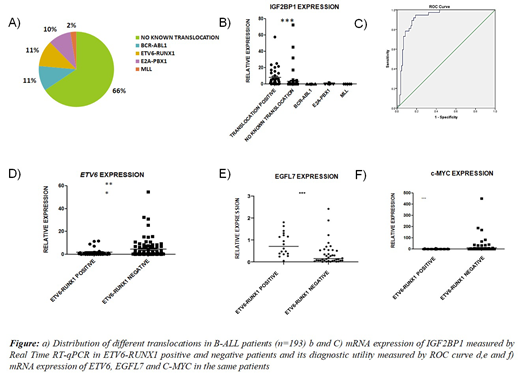
We collected 193 new paediatric pre B-ALL bone marrow samples. Most of the patients were under 5 years of age (73%) and only 3% of patients were more than 10 years. 66% of patients had no known translocation. Among the ones with translocation, ETV6-RUNX1 and BCR-ABL translocation were the commonest (11% each). 10% were E2A-PBX positive and only 3% had an MLL translocation. The translocation profile was very different from that of previously published reports.
E/R translocation generally portends a good prognosis. There was a significantly lower initial WBC count in E/R positive group when compared to the other groups. However, there was no significant difference in other blood parameters including initial Hemoglobin levels and platelet counts. Interestingly, the proportion of patients with a high number of blasts in peripheral blood on day 8 post chemotherapy was actually significantly higher in the E/R +ve group indicating that there might be a proportion of patients in the E/R +ve group with a worse prognosis. There was no difference in minimal residual disease between the E/R translocation positive and negative groups.
Real time RT-qPCR analysis of IGF2BP1 and ETV6 mRNA expression in bone marrow samples revealed significantly higher expression of IGF2BP1 in E/R positive B-ALL compared to E/R negative B-ALL samples. ETV6 mRNA relative expression on the other hand was found to be significantly lower in the E/R positive group. This corresponds to existing data that there is usually loss of the other wild type ETV6 allele in these patients.
We used data from multiple studies to narrow down on true targets of IGF2BP1 in B-ALL. We used a published eCLIP dataset of IGF2BP1 to identify cognate RNA targets. Over expression of these targets in E/R +ve samples was confirmed from the other previously published high throughput gene expression studies on B-ALL (MILE (microarray innovations in leukemia) study (n = 3242) and Palanichamy et al, 2016, J Clin Invest). We identified 13 such genes which were bound by IGF2BP1 and regulated by it in E/R +ve B-ALL. One of these targets, EGFL7, has previously been shown to play an important role in haematopoiesis and involved in Acute Myeloid Leukemia pathogenesis.
c-MYC is a known target of IGF2BP1 and. being a proto oncogene has an important role in B-cell proliferation. However, the biological and clinical role of cMYC and EGFL7 genes had not been evaluated in ETV6-RUNX1 positive Pre B-ALL.
Real time analysis of EGFL7 and cMYC expression in the patient bone marrow samples showed significantly altered expression in both groups. As expected, EGFL7 showed a significantly higher expression in E/R +ve samples. Unexpectedly, cMYC showed lower expression in E/R positive group. The expression of EGFL7 also showed a significant positive correlation with initial hemoglobin levels and a negative correlation with initial WBC counts. To study the effect of IGF2BP1 binding to the EGFL7 or c-MYC UTRs, they were cloned downstream of firefly luciferase and coexpressed with IGF2BP1 in HeLa cells. Interestingly, the dual luciferase assay revealed no stabilization of either of these UTRs by IGF2BP1. This reveals a much more complex regulation by IGF2BP1 of its targets which might involve RNA methylation.
To utilize the expression of IGF2BP1 and its target genes as a diagnostic marker for the presence of the E/R translocation, ROC curves using FISH/PCR as the gold standard test were prepared. The AUC (Area under curve) for ROC of the expression of IGF2BP1, c-MYC and EGFL7 were 0.917, 0.7 and 0.78 respectively. We also analyzed ROC curves using various combinations of these genes. Gene expression of IGF2BP1 alone was the best indicator of the presence of the ETV6-RUNX1 translocation with the highest sensitivity and specificity. In conclusion, relative expression of IGF2BP1 can serve as an excellent diagnostic marker for the E/R translocation with more than 90% sensitivity and 85% specificity.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal