Background
The acute leukemia of ambiguous lineage (ALAL) is high risk and uncommon in acute leukemia, including acute undifferentiated leukemia (AUL) and mixed phenotype acute leukemia (MPAL). On account of the rareness and age-influenced genome landscape, the genetic characterization was always limited and showed discrepancy among different cooperative groups.
Methods
By integrating the immunophenotype, cytogenetics, and molecular biology of ALAL patients, we found that the molecular profile played an important role in the diagnosis, treatment, and prognosis of ALAL patients. Through an in-depth analysis of the molecular features, we found the genetic differences between different subtypes of ALAL, and studied the unique gene mutation patterns and synergy of genomic abnormalities in different subtypes.
Results
Most patients in the cohort were children and young adults, and among B/myeloid and T/myeloid MPAL, while rare in AUL. Here we showed that ALAL patients show both genetic commonality and discrepancy among subtypes. Except for the BCR-ABL1 and KMT2A-r, which were mainly in B/myeloid MPAL, we reported that SET-NUP214 was recurrent in T/myeloid MPAL and AUL patients. The PHF6 were also frequently mutated in T/myeloid MPAL with most loss of function and significantly associated with SET-NUP214. Otherwise, there was a big difference between B/myeloid and T/myeloid MPAL in genetic profile. The most mutated genes in B/myeloid MPAL were CEBPA, RUNX1, NRAS, TET2, WT1, PTPN11, and FLT3, while NOTCH1, WT1, RUNX1, FLT3, PHF6, ETV6, and EZH2 were frequently in T/myeloid MPAL. Moreover, the alterations in the genes encoding transcriptional regulation were both recurrently mutated in B/myeloid and T/myeloid MPAL, especially WT1 and RUNX1, which were the most mutated genes and mutually exclusive with each other. Alterations in the genes involved in JAK_STAT signaling, RAS signaling, and epigenetic regulation were also recurrently mutated in B/myeloid and T/myeloid MPAL, despite some differences in incidence rate. However, the deletion of IKZF1 exclusively occurred in B/myeloid MPAL, while genes involved in NOTCH signaling were mainly altered in T/myeloid MPAL.
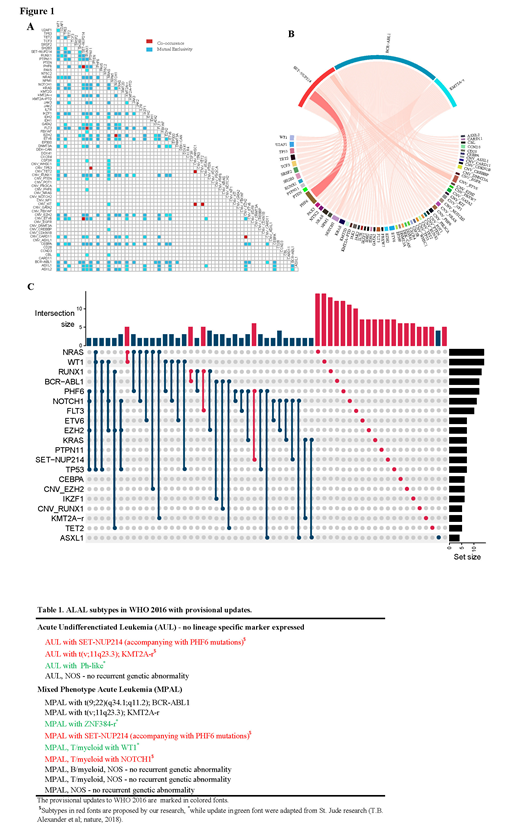
When analyzing the synergy among genetic abnormalities, we found that SET-NUP214 and PHF6 (p = 0.00002), FLT3 and RUNX1 (p = 0.009), BCR-ABL1 and RUNX1 (p = 0.02) were the most significantly co-occurred genes (Figure 1-A, B). Although the association between NRAS and WT1 were not significantly (p = 0.11), they co-occurred in five patients (Figure 1-C).
Since this was the first time to report that SET-NUP214 was recurrent in ALAL patients, we retrospectively review the therapy of patients with the aberration. According to the treatment of these ALAL patients, we confirmed that even with transplantation therapy the prognosis of SET-NUP214-positive ALAL patients was still very poor with a high risk of relapse.
Discussion
We charted the genomic landscape of ALAL to comprehensively reveal the commonality and discrepancy among subtypes of ALAL patients. The founding lesion of SET-NUP214 in early primitive hematopoietic cells was showed to be involved in the block of differentiation. Together with the loss of function in PHF6, which was showed a contribution to the lineage promiscuity and enhance the proliferation of hematopoietic cells, the combination event could make leukemia with ambiguous lineage. After deciphering the molecular characterization of ALAL patients, we proposed several candidate updates to WHO 2016 (Table 1), as these molecular features may help the diagnosis, therapy, and prognosis of the ALAL patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal