Background: In addition to acquired genetic changes in neoplastic plasma cells, progression of MGUS to multiple myeloma (MM) is mediated by changes in the proportion of dominant subclones whose expansion is driven by their interaction with the immune microenvironment. Experiments in mouse models of MM have identified a significant contribution to the microenvironment mediated via the gut microbiome and IL-17 responses (Calcinotto et al. Nat Commun 2018;(9)4832:1-7). Despite these preclinical studies, little is known about the composition of the human microbiome in MM at diagnosis, and we hypothesize that it is abnormal and can serve as an axis for immune dysregulation and selection of clonal B cells. In this study we analyzed gut microbial diversity in patients with MGUS and MM and compared these to healthy individuals.
Methods: Stool samples were obtained from 10 untreated MGUS and 10 MM patients with ECOG performance status 0-2. Exclusion criteria included those with prior anti-myeloma therapy, primary gastrointestinal disorders, or transient gastrointestinal processes or antibiotic use within the past 3 months. Healthy volunteers from the same geographic region served as controls. 16s rRNA gene sequencing was performed on fecal samples obtained with uBiome Explorer kits processed using Illumina NextSeq500. Qualitative analyses included determination of genera-level operational taxonomic units compared to an established healthy reference range and quantitative assessments of microbial diversity. Cluster analysis was performed using ClustVis (http://biit.cs.ut.ee/clustvis).
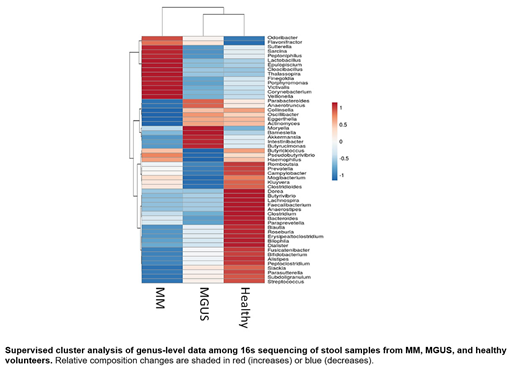
Results: The mean age at sampling was 74 years for MGUS and 71 for MM. Revised ISS stages I, II, and III for MM patients were 14%, 44%, and 42%, respectively. Median microbial diversity indices showed 84% in MM (range 38-96%), 85% in MGUS (range 42-93%), and 50% in healthy controls (range 23-89%). Supervised clustering of genus-level microbial sequences distinguished specific microbiota separating MM patients from MGUS and healthy controls (Figure). Odoribacter and Lactobacillus were the most enriched genera in MM compared to healthy, conversely Blautia and Faecalibacterium were most reduced. When comparing MM to MGUS, Kluyvera and Bacteroides were found to be most highly represented in MM, whereas Blautia and Parabacteroides showed the greatest decrease compared to MGUS. Data from a MM patient initially enrolled at diagnosis and resampled 4 months after achieving a partial response to induction with VCD showed a decrease in enrichment of Odoribacter from 12.2 to 2.1% as well as Bacteroides from 21.8 to 10.3%.
Conclusions: This study identifies distinct changes in the intestinal microbiota that distinguish individuals with MM from its precursor and healthy controls. This is the first time such changes have been identified in untreated MM patients, and some of the genera identified here, such as Faecalibacteria, have been shown to be similarly altered in recent reports of MM patients with persistently active disease despite treatment (Pianko et al. Blood Advances 2019;(3)13:2040-2044). These preliminary studies reveal distinct genus-level differences that exist between MM and MGUS, suggesting increasing dysbiosis with progression to MGUS and MM. We are currently extending these studies to include sequential sampling of additional patients following response to induction chemotherapy. These results provide a rationale for further studying the pathogenic impact of the intestinal microbiome on dysregulation of the MM microenvironment.
Morgan:Amgen, Roche, Abbvie, Takeda, Celgene, Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees; Celgene: Other: research grant, Research Funding. Braunstein:Celgene: Consultancy, Other: Advisory Boards; Amgen: Consultancy, Other: Advisory Board; AstraZeneca: Consultancy, Other: Advisory Board; Janssen: Consultancy, Other: Advisory Board, Research Funding; Verastem: Consultancy, Other: Advisory Board; Takeda: Consultancy, Other: Advisory Boards.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal