Background:The PML;RARA fusion protein is the hallmark driver of Acute Promyelocytic Leukemia (APL). The fusion disrupts retinoic acid signalling, leading to the proliferation of myeloid precursors halted at the promyelocyte stage of maturation. Chromatin conformation plays a fundamental role in controlling regulatory networks driving cell differentiation, however the genome wide organisational changes that occur during the PML;RARA differentiation block remains to be elucidated.
Aims: We have aimed to characterise three hallmarks of PML;RARA driven transcriptional mis-regulation: transcription factor binding, epigenetic remodelling and higher order chromatin organisation.
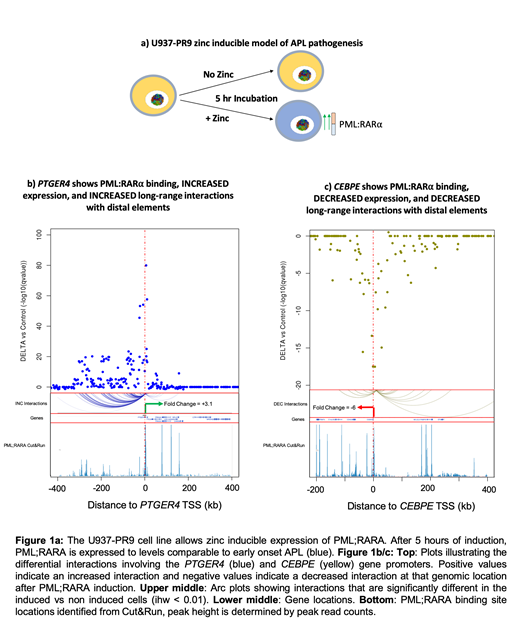
Methods: To a PML;RARA inducible U937-PR9 cell line model (Figure 1a), we have applied 1) Promoter Capture Hi-C (PCHi-C) to characterise long-range regulatory interactions, 2) Cut&Run, a newly described chromatin profiling technique, and 3) RNA-seq. All datasets are paired and have been modelled as induced vs non-induced.
Results: PML;RARA induction resulted in 855 significantly differentially expressed genes (DEGs, adjusted pvalue < 0.01). Up regulated genes (457) were enriched for genes involved in proliferative and oncogenic pathways. Down regulated genes (398) were enriched for drivers of cell differentiation, including the master regulators of myeloid differentiation: CEBPE, CEBPB and CEBPA. We identified 15,412 PML;RARA binding sites using Cut&Run (n=2). These sites encompassed 95% of previously uncovered PML;RARA binding sites, in addition to ~12,000 novel sites. 53% of these regions were distributed at gene promoters, 23% were intergenic and 24% at gene bodies. We observed a global decrease in H3k27ac after PML;RARA induction (86% regional loss), a significant proportion of these regions coincide with PML;RARA binding (Fisher's test 2.2-16). 1,900 PML;RARA peaks were associated with 66% DEGs. Interestingly, a significant proportion of PML;RARA peaks were not associated with DEGs. Applying PCHi-C (n=3) we identified >60,000 consistent differential interactions (DIs, ihw < 0.01), with a mean interaction distance of 100kb. 30,039 interactions increased and 30,403 interactions decreased after PML;RARA induction. Over half of DIs directly involved 8,066 PML;RARA binding sites (Fisher's test 2.2-16). We observed that genes losing long-range interactions were more likely to have decreased expression (59%) and this observation increased if a gene is also bound by PML;RARA (65%). SPI1, ID2, CEBPA, CEBPB, CEBPE, EGR1, and TFEB, key regulators of myeloid differentiation, display this pattern of PML;RARA driven negative regulation. We also observed that genes with increased expression were more likely to gain long-range interactions (60%). The top ranked gene across all datasets was the prostaglandin receptor 4 (PTGER4), a G-Protein coupled receptor highly expressed in AML and upregulated in our datasets (3.1 fold) (figure 1b). PTGER4 gained long-range interactions with an intergenic region ~300kb downstream of its promoter. This intergenic region is highly enriched for both H3k27ac and PML;RARA. This suggests that PML;RARA facilitates the long-range activation of PTGER4. We see a similar pattern to PTGER4 in 15% of DEGs where PML;RARA is anchored to both ends of the DI.
Summary/Conclusion: Applying novel NGS techniques to a simple model, we have highlighted that specific chromosome conformations are pivotal to the transcriptional mis-regulation driven by PML;RARA. We show PML;RARA may be directly involved in the re-organisation of the genome and this 3D architecture is pivotal in driving the Leukemia differentiation block.
Dillon:Abbvie: Consultancy, Honoraria; Novartis: Consultancy, Honoraria; Pfizer: Consultancy, Honoraria; TEVA: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal