Background
AML is a heterogeneous clonal disorder that is characterized by the accumulation of complex genomic alterations that affect disease biology and outcomes. Despite significant advances in our understanding of the impact of these mutations on overall survival (OS), established AML risk stratification guidelines are based primarily on cytogenetic analyses and a limited number of genes, don't take into account the complexity and the interaction between these mutations, and how particular constellations of genomic and clinical risk factors affect patient (pt) outcome.
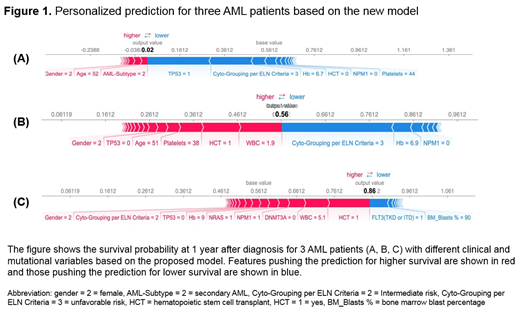
We developed a novel prognostic model that incorporates clinical, cytogenetic, and mutational data to determine personalized outcomes specific to a particular pt.
Method
A total of 792,779 genomic and clinical data points from 3,421 pts were analyzed. The cohort was comprised of five independent datasets: 443 pts from the Beat AML Master Trial (Tyner et al, Nature, 2018), 855 pts from Cleveland Clinic, 414 pts from Munich Leukemia Laboratory (MLL), 1,509 pts from the German-Austrian Study Group (Papaemmanuil et al, NEJM, 2016), and 200 pts from The Cancer Genome Atlas (NEJM, 2013). A panel of 44 gene mutations commonly implicated in AML was used in the analysis, along with numerous cytogenetic and clinical variables such age, white blood cell count WBC) at diagnosis, and AML subtype (primary vs. secondary vs. therapy-related. A machine learning algorithm capable of accounting for survival (XGBOOST) was used to build the new model, in which clinical and molecular variables were randomly selected for inclusion in determining OS. Feature extraction algorithms were used to isolate the most important variables that impacted decision making within the model. The algorithm can also plot the important features that are specific for a given pt and show the impact of each feature on the outcome (positive vs. negative). The C-index was used to evaluate the accuracy of the new model compared to 2017 ELN risk classification.
Results
The median age of the cohort was 56 years (range, 18-100); 1,122 pts (32.8%) had favorable risk cytogenetics per ELN criteria, 956 (27.9%) intermediate (INT), and 1,343 (39.3%) adverse. The most commonly mutated genes were: NPM1 (24%), FLT3 (23%), DNMT3A (20%), NRAS (13%), IDH2 (11%), RUNX1 (10%) and TET2 (10%). Mutations occurred in different frequencies in each cytogenetic risk group. The most commonly mutated genes in the favorable risk group were: NRAS (30%), KIT (23%), FLT3 (17%), and KRAS (8%). The most commonly mutated genes in the INT risk group were: NPM1 (28%), FLT3 (26%), DNMT3A (22%), IDH2 (12%), TET2 (11%), NRAS (11%), and RUNX1 (11%). The most commonly mutated genes in pts with adverse cytogenetics included: TP53 (34%), DNMT3A (13%), NRAS (11%), RUNX1 (10%), PTPN11 (8%), IDH2 (7%), U2AF1 (6%) and FLT3 (6%). All genomic-clinical variables were included in the machine learning algorithm. Variable importance analyses (the most important variables that contributed to the outcome) and multiple backward elimination analyses (identifying the least number of variables that can provide the least error rate) identified the following variables that impacted OS: age, transplant (yes vs. no), WBC, bone marrow blast %, cytogenetics, ASXL1, CEBPA, DNMT3A, FLT3, KDM6A, KIT, KRAS, NPM1, NRAS, PHF6, PTPN11, RUNX1, TET2, and TP53. The clinical and mutational variables that impacted each pt outcome can be visualized in a highly personalized manner, Figure 1.
The C-index for the new model was 0.80 which significantly outperformed ELN classification (0.59). When applying the new model to each of the five patient cohorts, the c-indices remained high and were as follows: Beat AML (0.81), Cleveland Clinic (0.85), MLL (0.83), Papaemmanuil E, et al (0.79), and TCGA (0.80).
Conclusions
Genomic alterations have a differential impact on OS in each cytogenetic risk group, highlighting the complexity of incorporating these mutations into risk stratification. A personalized prediction model based on clinical-genomic data can accurately provide survival unique to each individual pt and can significantly outperform ELN classifications or any currently available models. To ease the translation of this model into the clinic, a web application is currently under development and will be publicly available for use.
Meggendorfer:MLL Munich Leukemia Laboratory: Employment. Mukherjee:Bristol-Myers Squibb: Speakers Bureau; Takeda: Membership on an entity's Board of Directors or advisory committees; Pfizer: Honoraria; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Projects in Knowledge: Honoraria; Celgene Corporation: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Partnership for Health Analytic Research, LLC (PHAR, LLC): Consultancy; McGraw Hill Hematology Oncology Board Review: Other: Editor. Walter:MLL Munich Leukemia Laboratory: Employment. Hutter:MLL Munich Leukemia Laboratory: Employment. Maciejewski:Novartis: Consultancy; Alexion: Consultancy. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Sekeres:Millenium: Membership on an entity's Board of Directors or advisory committees; Syros: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Nazha:Jazz Pharmacutical: Research Funding; Novartis: Speakers Bureau; Incyte: Speakers Bureau; Tolero, Karyopharma: Honoraria; Abbvie: Consultancy; Daiichi Sankyo: Consultancy; MEI: Other: Data monitoring Committee.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal