Background: Chronic myeloid neoplasms are heterogeneous malignancies caused by sequential accumulation of genetic lesions in hematopoietic stem cells with a tendency to evolve towards acute myeloid leukemia. Genomic approaches can stratify patients according to their mutational landscape but are limited in predicting response to therapeutic agents. Reliable biomarkers to identify patients' chances for response to targeting compounds remain a crucial necessity in the current post-genomic era of precision medicine. We hypothesize that differential phosphoproteomic (PP) profiles might represent a suitable functional biological layer that allows to identify relevant determinants of oncogenic phenotypes.
Aim: The aim of the project was to build a bioinformatics pipeline using PP data to i) identify differentially phosphorylated sites, ii) infer targetable kinases and iii) characterize involved oncogenic pathways. Here, we present results from further exploratory experiments of a previously established PP analysis pipeline that enables to infer sensitive/bypassing kinase activities in a Midostaurin/PKC412 (MIDO) resistant myeloid cell line model.
Methods: For the validation of our analysis pipeline, we have previously used the human myeloid cell lines K562 and MOLM13, driven by the oncogenic BCR-ABL1 and FLT3 kinases, and exposed to the kinase inhibitors Nilotinib (NILO) and MIDO, respectively. For the current biological study, we applied our pipeline to explore MIDO resistance mechanisms in published MIDO-resistant (rMOLM13) and sensitive (sMOLM13) cell lines (kindly provided by E. Weisberg, Dana Farber Cancer Institute, USA). r/sMOLM13 were cultured in triplicates for 24hrs and subsequently exposed for 1h to 25nM MIDO or DMSO (CTRL). PPs were enriched with titanium-dioxide and analyzed by mass spectrometry (nanoLC-MS2). Our previously validated Kinase Activity Enrichment Analysis (KAEA) bioinformatics pipeline was further applied to infer kinase activities based on the SetRank enrichment algorithm. KAEA integrates substrate-kinase datasets from five experimentally validated databases complemented with NetworKIN in-silico predictions. The pipeline is supported by a Shiny web-app interface to allow interactive visualization and interrogation of the data.
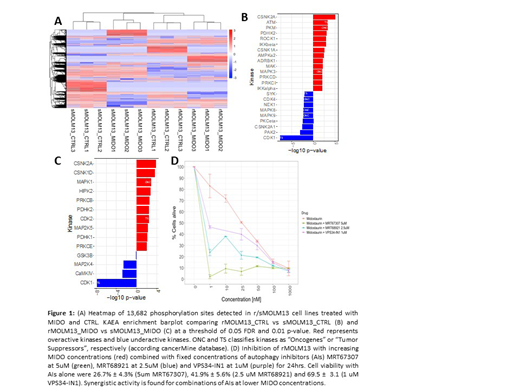
Results: In our previous validation experiments, K562 treated with NILO showed expected inhibition of ABL1 and KIT, whereas MOLM13 treated with MIDO showed expected inhibition of PRKC and downstream kinases of FLT3 (AKT1 and MAPK). In the current biological exploration, we compared rMOLM13 vs sMOLM13 exposed to MIDO and CTRL. rMOLM13 showed slower proliferation and vacuolization compared to sMOLM13. 13,682 phosphorylation sites were identified and the three replicates clustered adequately in the corresponding four conditions sMOLM13_CTRL, sMOLM13_MIDO, rMOLM13_CTRL and rMOLM13_MIDO (fig. 1A). Clusters of over- or underexpressed sites associated with distinct conditions emerged in the heatmap. We performed a KAEA comparison of rMOLM13_MIDO vs sMOLM13_MIDO (fig. 1B) and rMOLM13_CTRL vs sMOLM13_CTRL (fig. 1C). CDK1 and other CDKs enriched as underactive in rMOLM13, in line with the observed reduced proliferation. Most interestingly, kinases PDHK1, PDHK2 and PKM, all pyruvate kinases involved in regulation of mitochondrial metabolism, enriched as overactive, as well as MAPK1/3 and CK2. Reduced proliferation, vacuolization and patterns of enriched kinases pointed towards autophagy as potential resistance mechanism in rMOLM13. We performed preliminary experiments with autophagy inhibitors in combination with MIDO, which supported a synergistic activity (fig. 1D). More results of combined inhibition of the identified kinases will be shown at the meeting.
Conclusions: With our PP analysis pipeline, we were able to characterize, in unanticipated detail, dynamic changes of PP profiles, kinase activities and potentially involved oncogenic pathways in a MIDO resistant myeloid cell line model. We identified biologically relevant mechanisms of resistance that will be explored as potential targets for combination therapy. Our experiments in myeloid cell lines provide a proof of concept for our analysis pipeline. They allow us to move forward to the analysis of primary patient samples and promote precision medicine based on functional biomarkers identified by PP in patients with myeloid neoplasms.
Jankovic:Celgene: Other: financial support for travel. Bonadies:Roche: Other: financial support for travel, Research Funding; Novartis: Other: financial support for travel, Research Funding; Celgene: Other: financial support for travel, Research Funding; Janssen: Other: financial support for travel; Amgen: Other: financial support for travel; Sanofi Genzyme: Other: financial support for travel.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal