Current risk stratification for newly diagnosed patients with chronic myelomonocytic leukemia (CMML) includes clinical and genetic features accounting for its variable disease course. Allogeneic stem cell transplantation still remains the only curative treatment option, and prognostication of posttransplant outcome may be improved using molecular information. Here, we aim to evaluate the molecular profile and its role on posttransplant outcome in a multicenter CMML cohort.
Mutation analysis was performed on DNA from bone marrow mononuclear cells or peripheral granulocytes collected prior to transplant and included previously published CMML-associated genes (i.a. SETBP1, ASXL1, RUNX1, NRAS, KRAS, TET2, CBL, IDH1/2, SF3B1, DNMT3A, EZH2, ZRSR2, U2AF1). Current prognostic models were calculated at time of transplant. Patients with transformation to acute leukemia were excluded. Top predictors of posttransplant outcome were identified using the Random Forest algorithm. Main end points were overall survival (OS) and non-relapse mortality (NRM).
The total cohort consisted of 185 patients of whom seven had CMML-0, 100 CMML-1, and 78 CMML-2 at time of transplant. The median follow-up was 74 months and 6-year OS was 37% for the total cohort and differed for CMML-0 (57%), CMML-1 (43%), and CMML-2 (29%). Relapse and NRM were 27% and 44% for the total cohort being 17% and 31% for CMML-0, 23% and 40% for CMML-1, and 34% and 40% for CMML-2. Most frequently mutated genes were: TET2 (55%), ASXL1 (41%), SF3B1 (38%), DNMT3A (27%), ZRSR2 (22%), NRAS (21%), EZH2 (21%), RUNX1 (17%), and SETBP1 (17%). Ninety-two percent of patients showed at least one somatic mutation. More than three mutations were present in 49% of all patients and in 29% of CMML-0, 50% of CMML-1, and 49% of CMML-2 patients. Frequencies according to CMML-specific prognostic scoring system (CPSS) and its molecular refinement (CPSS-mol) were 8% and 6% (low risk), 31% and 18% (intermediate-1 risk), 43% and 40% (intermediate-2 risk), and 18% and 36% (high risk). Transplants were received from matched unrelated (51%), mismatched unrelated (25%), matched related (21%), or mismatched related donors (3%). Conditioning intensity was reduced (49%), myeloablative (43%), or non-myeloablative (8%). Median age of patients was 60 years, 29% were female, 30% had a Karnofsky performance status <90%, and 15% had a comorbidity index >3.
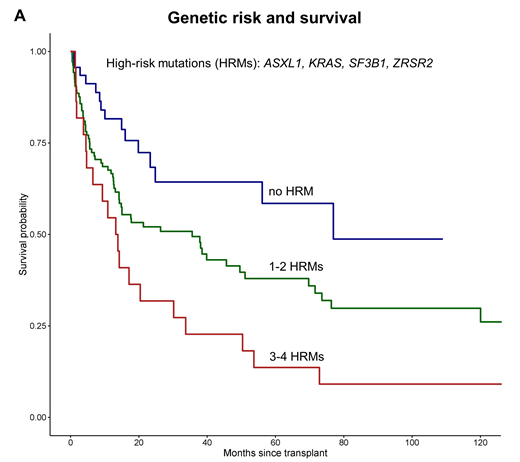
In the first step of the OS analysis, the algorithm identified mutations in ASXL1, KRAS, SF3B1, ZRSR2 as high-risk mutations (HRM) predicting worse OS. In addition, the number of the HRMs was associated with worse OS. In the next step, the algorithm automatically stratified this information into three distinct risk groups: the absence of HRMs (reference; low risk), presence of 1-2 HRMs (HR, 1.81; intermediate-risk), and 3-4 HRMs (HR, 2.93; high-risk). Corresponding 6-year OS was 59% for the low-risk, 34% for the intermediate-risk, and 14% for the high-risk group (P<.001; Figure 1A). Furthermore, the absence of HRMs was associated with lower NRM (15%) compared with present HRMs (46%; P=.01). In contrast, the CPSS-mol genetic risk classification including ASXL1, RUNX1, NRAS, and SETBP1 mutations showed no distinct 6-year OS or NRM (P=.15, respectively).
Next, we adjusted the impact on OS of the proposed genetic risk for other factors included in the CPSS-mol. Higher genetic risk was independently associated with increased hazard for death (with the low-risk group as reference) showing HRs of 1.70 for the intermediate-risk and 2.83 for the high-risk group (P<.001). This model showed a concordance index of 0.633 versus CPSS-mol (0.597) or the CPSS (0.572) suggesting utility of transplant-specific prognostication.
Therefore, we evaluated the multivariable effect on posttransplant outcome including the following independent clinical and molecular predictors: genetic risk, % of peripheral and bone marrow blasts, leukocyte count, and performance status. This model was predictive of OS and NRM (P<.001, respectively), and showed increased prognostic precision for OS, reflected in a concordance index of 0.684.
In conclusion, mutations in ASXL1, KRAS, SF3B1, ZRSR2, and the number of these mutations predict OS and NRM in CMML undergoing transplantation. Accounting for these genetic lesions may improve the prognostic precision and patient counseling in the transplant setting.
Bogdanov:Jazz Pharmaceuticals, MSD.: Other: Travel subsidies. Stoelzel:Neovii: Other: Travel funding; JAZZ Pharmaceuticals: Consultancy; Shire: Consultancy, Other: Travel funding. Rautenberg:Jazz Pharmaceuticals: Other: Travel Support; Celgene: Honoraria, Other: Travel Support. Dreger:Neovii, Riemser: Research Funding; MSD: Membership on an entity's Board of Directors or advisory committees, Other: Sponsoring of Symposia; AbbVie, AstraZeneca, Gilead, Janssen, Novartis, Riemser, Roche: Consultancy; AbbVie, Gilead, Novartis, Riemser, Roche: Speakers Bureau. Finke:Riemser: Honoraria, Other: research support, Speakers Bureau; Neovii: Honoraria, Other: research support, Speakers Bureau; Medac: Honoraria, Other: research support, Speakers Bureau. Kobbe:Pfizer: Honoraria, Other: Travel support; Takeda: Honoraria, Other: Travel support; Celgene: Honoraria, Other: Travel support, Research Funding; Jazz: Honoraria, Other: Travel support; Amgen: Honoraria, Other: Travel support, Research Funding; Biotest: Honoraria, Other: Travel support; MSD: Honoraria, Other: Travel support; Neovii: Honoraria, Other: Travel support; Abbvie: Honoraria, Other: Travel support; Novartis: Honoraria, Other: Travel support; Roche: Honoraria, Other: Travel support; Medac: Honoraria, Other: Travel support. Platzbecker:Celgene: Consultancy, Honoraria, Research Funding; Abbvie: Consultancy, Honoraria; Novartis: Consultancy, Honoraria, Research Funding. Robin:Novartis Neovii: Research Funding. Beelen:Medac GmbH Wedel Germany: Consultancy, Honoraria. Kroeger:JAZZ: Honoraria; Neovii: Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Sanofi-Aventis: Honoraria; Novartis: Honoraria, Research Funding; Medac: Honoraria; DKMS: Research Funding; Riemser: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal