Background: Imatinib resistance mutation analysis (IRMA) or abl kinase domain mutation analysis is performed in patients with Chronic myeloid leukemia (CML) whenever the response to treatment is inadequate. We have analyzed the reports of IRMA at our centre.
Methods: The clinical details of 71 patients with CML on Imatinib, who underwent IRMA testing during the period of January 2017 to March 2019 were collected from the patient records and analyzed. IRMA was performed for failure or warning or progression, anytime during the course of treatment. IRMA was done by either Sanger sequencing (n=45) or next generation sequencing (n=26, Illumina, NGS). The associations between variables were tested using Chi - Square test.
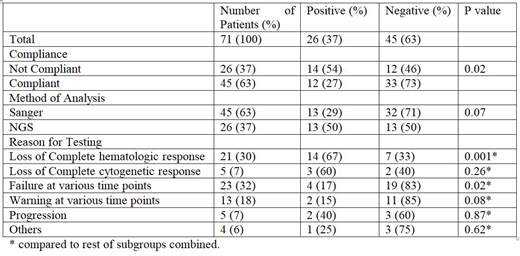
Results: Median age at diagnosis of 71 patients was 44 years (Range 18 - 71 years). Males constituted 70% (n=50). At diagnosis, 92% (n=65) of patients were in chronic phase and the remainder were in accelerated phase (n=4) or blast crisis (n=2). Mutations in the abl kinase domain were detected in 26 patients (37%). Next Generation Sequencing (NGS) could identify more mutations (13/26 - 50%) compared to conventional Sanger Sequencing (13/45 - 29%), but the difference was not significant (p=0.07). NGS could identify three or more mutations in 5 patients in contrast to Sanger. All the mutations detected were those previously described except for an insertion of 35bp near the C-Terminal which was identified in 3 patients. E459K translocation was identified in 6 patients. E355G translocation was identified in 4 patients. F359V, M351T, Y253H, G250E, H396R, T315I translocations were identified in 3 patients each. Patients who were not compliant to therapy had increased frequency of mutations (14/26 - 54%) compared to those who were compliant (12/45 - 27%), which was significantly different (p=0.02). Patients who had loss of complete hematological response (CHR) had significantly higher frequency of mutations (14/21- 67%) compared to other reasons for performing the test (p=0.001). Patients who had failure to achieve targets at various time points had a significantly lower frequency of mutations (4/23 - 17%, p=0.02) compared to other reasons for performing the test.
Conclusion: Patients who were not compliant for treatment were more likely to have mutations. Loss of CHR showed an increased frequency compared to other reasons. NGS could identify mutations in more number of patients. NGS identified numerically higher mutations in patients. Larger prospective data are needed to confirm these observations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal