It is increasingly evident that the composition of the tumor microenvironment (TME) and the interplay between malignant and immune cells determine the efficacy of antitumor immune responses, whether pre-existing or induced by therapy. To profile the complexity and plasticity of the TME and track antitumor immunity, we performed single cell RNA sequencing on tumor fine needle aspirates and peripheral blood of lymphoma patients undergoing immunotherapy on a clinical trial (NCT02927964).
In this in situ vaccination study motivated by compelling preclinical data (Sagiv-Barfi et al., Blood 2015), patients with low-grade lymphoma receive local low-dose radiation and a TLR9 agonist (CpG) intratumorally to one site of disease. In the second week, after the second of 5 intratumoral CpG injections, they begin taking oral ibrutinib. Tumor fine needle aspirates (FNAs)and peripheral blood samples are obtained prior to treatment, and at 1 week and 6 weeks after treatment start. Single cell preparations of FNA and blood samples are processed using the 10X Genomics platform (based on Zheng et al., Nat Commun 2017). Libraries are sequenced to an average targeted depth of 50,000 reads/cell. Identification of variable genes, principal component and graph-based clustering, and differential expression analysis of single-cell gene expression data is performed using the Seurat algorithm (Butler et al., Nat Biotech, 2018).
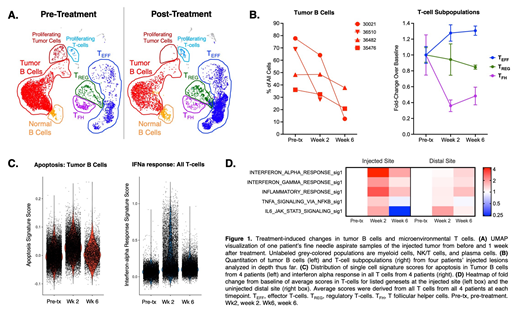
We have prepared sequencing libraries from 58 tumor FNA and peripheral blood samples from 7 patients thus far. For the first 6 patients, approximately 3,000-10,000 cells per sample have been sequenced, with excellent sequencing quality metrics. Evaluating treatment-induced changes at the CpG-injected site across patients, we found a loss of tumor B cells and T follicular helper cells (Tfh) along with increases in CD8 and CD4 effector cells. Genes associated with cell death, DNA damage response, interferon response, and antigen presentation were induced in tumor B cells after treatment, while genes associated with several signaling pathways, including Myc, Wnt/beta-catenin, and MTOR were down-regulated. In contrast, treatment-induced changes in the tumor-resident T-cells were dominated by a strong interferon response (to both interferon-alpha and -gamma), strongest in CD4 effector and memory populations and weakest in T follicular helper cells and exhausted T cells. T-cells also showed induction of genes associated with TNF-alpha signaling, IL6 signaling, and inflammatory chemokines and cytokines. Interestingly, at the distal, non-injected site, similar changes occurred with smaller magnitude and more so at the week 6 timepoint. Further expression analysis, sequencing and analysis of single cell TCR clonotypes, and relation of results to clinical data is ongoing.
Deep profiling of serial biopsies offers a comprehensive view of the evolving immune microenvironment during immunotherapy. By sampling multiple tumors over time in patients undergoing in situ vaccination, we identify significant and distinct transcriptional shifts in different tumor microenvironmental subpopulations at both the injected and un-injected sites, including activation of T-cells both sites with different dynamics. Our approach represents successful application of single-cell genomics to a clinical trial, illuminating cellular dynamics induced by treatment.
Levy:Apexigen: Membership on an entity's Board of Directors or advisory committees; Nohla: Membership on an entity's Board of Directors or advisory committees; Spotlight: Membership on an entity's Board of Directors or advisory committees; 47 Inc: Membership on an entity's Board of Directors or advisory committees; XCella: Membership on an entity's Board of Directors or advisory committees; Immunocore: Membership on an entity's Board of Directors or advisory committees; Walking Fish: Membership on an entity's Board of Directors or advisory committees; Five Prime: Membership on an entity's Board of Directors or advisory committees; Corvus: Membership on an entity's Board of Directors or advisory committees; Quadriga: Membership on an entity's Board of Directors or advisory committees; BeiGene: Membership on an entity's Board of Directors or advisory committees; GigaGen: Membership on an entity's Board of Directors or advisory committees; Teneobio: Membership on an entity's Board of Directors or advisory committees; Sutro: Membership on an entity's Board of Directors or advisory committees; Checkmate: Membership on an entity's Board of Directors or advisory committees; Nurix: Membership on an entity's Board of Directors or advisory committees; Dragonfly: Membership on an entity's Board of Directors or advisory committees; Innate Pharma: Membership on an entity's Board of Directors or advisory committees; Abpro: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal