Background: Core-binding factor acute myeloid leukemias (CBF-AML) are characterized by inv(16)/t(16;16)(p13.1;q22) or t(8;21)(q22;q22.1) and encompass a distinct subgroup of AML with a favorable prognosis. This subgroup represents approximately 15% of newly diagnosed adult AML and is more frequent in young adults. With intensive chemotherapy, nearly all patients with CBF-AML achieve complete remission (CR) and the long-term overall survival (OS) is 75 to 85%. However, up to 40% of adult patients will relapse after achieving CR and some patients may die of progressive disease. Early identification of patients with higher risk disease would help to select patients who may benefit from targeted or higher intensity therapeutic approaches to reduce their risk of relapse. The aim of this study was to develop a gene expression signature to predict at time of diagnosis the risk of relapse in patients with CBF-AML.
Methods: We analyzed the RNA sequencing (RNA-Seq) data of 44 diagnostic specimens from patients with de novo CBF-AML treated with intensive chemotherapy (7+3 regimen followed by high-dose cytarabine consolidations). We performed logarithmic transformation and standardization of RNA-Seq data normalized in RPKM and excluded genes with very low expression (< 1 RPKM in all specimens). We selected the genes which were significantly associated with relapse-free survival (RFS) in univariate Cox proportional hazard (CPH) models with a significance level of < 0.05. In this subset of genes, we fitted a CPH regression model with the LASSO algorithm using L1 penalty regularization on coefficients to minimize the 10-fold cross-validation partial likelihood deviance. We derived a score from the genes selected by the LASSO algorithm based on the sum of each gene expression weighted by their regression coefficients. The prognostic impact of the score was evaluated in CPH regression models for RFS and OS. We also assessed the prognostic impact of individual genes included in the score after dichotomizing them into binary variables.
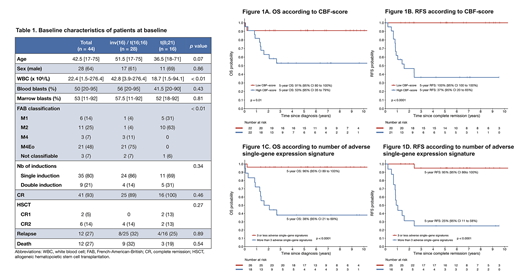
Results: Our cohort of CBF-AML included 28 patients with inv(16)/t(16;16) and 16 patients with t(8;21). Characteristics of the patients at diagnosis are summarized in table 1. The median age at diagnosis was 42.5 year-old (range, 17-75). Patients with inv(16)/t(16;16) had a higher white blood cell count at diagnosis (median 42.8 vs 18.7, p < 0.01). Rates of CR were 100% (16/16 pts) and 89% (25/28 pts) for patients with t(8;21) and inv(16)/t(16;16), respectively. The 3 pts with inv(16)/t(16;16) who did not achieve CR were older than 60 year-old and died during induction of chemotherapy-related complications. With a median follow-up of 6.8 years, 28 patients remain alive in remission, 12 patients relapsed and 1 patient died in CR. The 5-year OS and RFS are 72% (95% confidence interval [CI], 60 - 87%) and 67% (95% CI, 54 - 84%), respectively.
Among 24 586 genes for which RNA-Seq data was available, 506 genes (2.06%) were significantly associated with RFS in univariate analyses. Nine genes were selected by the LASSO algorithm and were used for calculation of the CBF-score: CPSF6, DUSP2, H3F3A, NAB2, NOTCH3, PAG1, RASGEF1A, THRAP3, TRIM24. The CBF-score as continuous variable was significantly associated with RFS (p < 0.001) and OS (p = 0.003), and remained significant for both outcomes when adjusted for age. When the CBF-score was dichotomized on the median, none of the patients with a low score relapsed whereas 12/21 (57.1%) patients with a high score relapsed. The estimated 5-year RFS rates were 100% (95% CI, 100 - 100%) and 37% (95% CI, 20 - 65%) in patients with a low and high CBF-score, respectively (p < 0.001) (Figure 1A-1B). When each gene of the score were individually dichotomized, all except NOTCH3 were significantly associated with worse RFS. For any additional adverse single-gene expression signature, the hazard ratio for RFS was 2.17 (p < 0.001). In patients with 4 or more adverse single-gene signatures, the 5-year RFS was 25% (95% CI, 11 - 58%) versus 95% (95% CI, 86 - 100%) in patients with 3 or less single-gene signatures (Figure 1D).
Conclusion: We developed in silico a 9-gene expression signature that predicts with high accuracy patients with CBF-AML at high risk of relapse (high CBF-score) and patients who are likely to be cured with standard chemotherapy (low CBF-score). This prognostic signature is currently being validated in independent internal and external validation cohorts.
Sauvageau:ExCellThera: Consultancy, Employment, Equity Ownership, Membership on an entity's Board of Directors or advisory committees, Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal