In this issue of Blood, Downes et al provide an updated report on the use of a high-throughput screening (HTS) panel developed by the ThromboGenomics group in the United Kingdom for genetic analysis of patients with coagulation, platelet, or thrombotic disorders.1
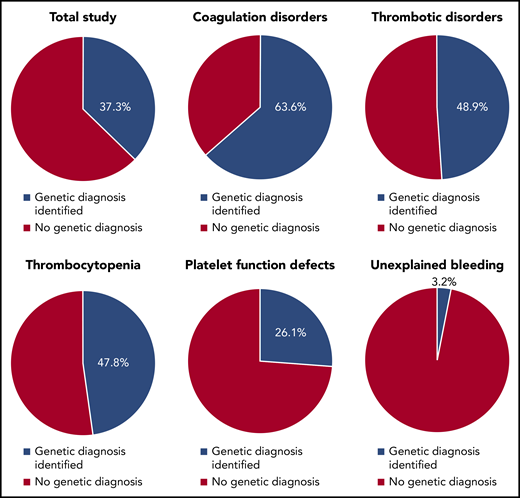
Diagnostic yield of the ThromboGenomics 96-gene HTS panel in patients with bleeding, platelet, or thrombotic disorders.
Diagnostic yield of the ThromboGenomics 96-gene HTS panel in patients with bleeding, platelet, or thrombotic disorders.
Clinical hematologists across the globe find that unexplained bleeding or clotting disorders make up a substantial part of new patient evaluations. In the 1980s, the genes encoding factor VIII, factor IX, and von Willebrand factor were characterized, which opened the door to genetic analysis at dedicated research centers. As a result, pathogenic gene variants were identified in about 95% of cases of hemophilia A or B and about two-thirds of those with type 1 von Willebrand disease.2,3 A short time later, various inherited thrombophilias were also described, leading to the demonstration of factor V Leiden, prothrombin gene mutation, antithrombin deficiency, protein C deficiency, or protein S deficiency in about 10% to 20% of patients with venous thromboembolism.4 In 2004, the World Health Organization created international standards for common genetic tests, beginning with factor V Leiden, reflecting the high demand for clinical thrombophilia testing around the world.
Despite these advances, the vast majority of patients with thrombotic or bleeding disorders, including platelet function defects, do not have one of the above genotypes. Until recently, a genetic diagnosis has largely remained elusive for these patients.
In the last decade, the advent of next-generation sequencing (NGS) led to the discovery of many new genes with potential roles in hemostasis and platelet function. In 2016, a landmark study by the ThromboGenomics group described the use of a 63-gene HTS panel in 296 patients with bleeding, platelet, or thrombotic disorders, with encouraging results, although the study population was weighted toward individuals with platelet function defects.5,6 Inclusion of genes in the ThromboGenomics panel and interpretation of genetic test results were determined by a multidisciplinary team of laboratory and clinical specialists. The likelihood of establishing a genetic diagnosis was dependent on pretest probability as gauged by clinical laboratory testing. In this setting, other investigative groups, including our own, also developed expanded NGS panels for use in patients with bleeding or thrombotic disorders, all with promising findings.7-9
The updated study by Downes et al improves upon their prior work in several key ways. The number of genes in the HTS panel has now been expanded to 96, reflecting the discovery and validation of new genes involved in hemostasis and platelet function over the intervening years. Methodologic changes have vastly improved the detection of copy number and intronic variants. The panel has now been tested in 2390 patients, representing a broad distribution of hemostatic, platelet, and thrombotic disorders. The consensus criteria for establishing a genetic diagnosis as determined by the multidisciplinary team have been refined.
In their new study, Downes et al were able to identify a genetic diagnosis in 37.3% of all patients (see figure). Interestingly, the likelihood of establishing a genetic diagnosis was a function of disease phenotype. The highest diagnostic rate was seen in patients with coagulation disorders (63.6%), followed by thrombotic disorders (48.9%), thrombocytopenia (47.8%), and platelet function defects (26.1%). By contrast, very few patients with unexplained bleeding achieved a genetic diagnosis (3.2%).
What implications do these findings have in the clinic? The high diagnostic yield of the ThromboGenomics panel in patients with coagulation disorders is as interesting as the very poor diagnostic yield in those with unexplained bleeding. Although numerous patients and family members who undergo ThromboGenomics testing will now have the satisfaction of being able to define their diseases genetically, the fact that more than half of patients have a negative ThromboGenomics evaluation suggests either that other genes not included in the HTS panel may be involved or that epigenetic, proteomic, or nongenetic factors may have important roles in these conditions. Subjectivity in how patients and providers distinguish normal from abnormal bleeding may also underlie some of the low diagnostic rate in patients with unexplained bleeding. Among individuals in whom a genetic diagnosis is established by using the ThromboGenomics HTS panel, a direct impact on management is uncertain, although it is possible that further research may reveal certain genotype-phenotype associations akin to those described in hemophilia and von Willebrand disease.10
The availability of NGS testing for clinical purposes is dramatically changing our understanding of human diseases. The ThromboGenomics HTS panel represents the most comprehensive NGS test available to date for the clinical study of patients with hemostatic or platelet disorders. The findings from the ThromboGenomics study have the potential to transform how clinicians and providers evaluate and manage patients with coagulation, platelet, or thrombotic disorders while identifying gaps in our current knowledge that require further discovery.
Conflict-of-interest disclosure: J.M.C. served on the scientific advisory boards for Bristol-Myers Squibb and Portola, was a consultant for Bristol-Myers Squibb, and received research funding from CSL Behring. A.I.L. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal