Key Points
Genetic deletion of Gdf11 does not affect red blood cell formation during homeostasis or after transplant.
Hematopoietic stem cell function is preserved in mice lacking Gdf11 expression within the blood lineage.
Abstract
Tightly regulated production of mature blood cells is essential for health and survival in vertebrates and dependent on discrete populations of blood-forming (hematopoietic) stem and progenitor cells. Prior studies suggested that inhibition of growth differentiation factor 11 (GDF11) through soluble activin receptor type II (ActRII) ligand traps or neutralizing antibodies promotes erythroid precursor cell maturation and red blood cell formation in contexts of homeostasis and anemia. As Gdf11 is expressed by mature hematopoietic cells, and erythroid precursor cell expression of Gdf11 has been implicated in regulating erythropoiesis, we hypothesized that genetic disruption of Gdf11 in blood cells might perturb normal hematopoiesis or recovery from hematopoietic insult. Contrary to these predictions, we found that deletion of Gdf11 in the hematopoietic lineage in mice does not alter erythropoiesis or erythroid precursor cell frequency under normal conditions or during hematopoietic recovery after irradiation and transplantation. In addition, although hematopoietic cell-derived Gdf11 may contribute to the pool of circulating GDF11 protein during adult homeostasis, loss of Gdf11 specifically in the blood system does not impair hematopoietic stem cell function or induce overt pathological consequences. Taken together, these results reveal that hematopoietic cell–derived Gdf11 is largely dispensable for native and transplant-induced blood formation.
Introduction
Growth differentiation factor 11 (GDF11), a member of the activin subclass of transforming growth factor-β ligands, was recently implicated as a regulator of erythropoiesis in studies employing activin receptor type II (ActRII) ligand traps,1,2 generated by fusion of the human or mouse immunoglobulin G1 Fc domain to the extracellular domain of either ActRIIA (ACE-011 or RAP-011)1 or ActRIIB (ACE-536 or RAP-536).2 Administration of such chimeric receptors promotes erythropoiesis in multiple species and contexts; however, because these molecules inhibit signaling by multiple ligands (including activin A, activin B, GDF8, and GDF11), the conclusion that improved erythropoiesis occurs specifically due to inhibition of GDF11 requires further testing. Furthermore, whether hematopoietic cell–derived Gdf11 might play a role in the formation and/or function of additional blood lineages or precursor cells, or in the previously documented effects of GDF11 on aging phenotypes,3,4 has not been explored. Here, we address these outstanding questions using genetic loss-of-function mouse models and find that, contrary to prior suggestions,1,2 hematopoietic cell–derived Gdf11 is largely dispensable for steady-state and regenerative hematopoiesis (including erythropoiesis), and its deletion is insufficient to accelerate the emergence of aging phenotypes (see supplemental text, available on the Blood Web site).
Study design
Mice
Mouse genotypes, breeding schemes, and genotyping strategies are provided in supplemental Methods. Analyses used age-matched male and female mice, with no specific randomization or blinding protocols. All studies were approved by Harvard’s Institutional Animal Care and Use Committee.
RNA isolation, cDNA synthesis, 3′ RACE, and real-time PCR
Details of sample preparation and polymerase chain reaction (PCR) are provided in supplemental Methods. Primers are listed in supplemental Table 1.
Complete blood count analysis
Blood was collected in EDTA-coated Microtainer tubes (BD). Complete blood counts were analyzed using a VetScan HM5 instrument (Abaxis).
Serum collection and LC-MS/MS analysis
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis was performed by the Brigham Research Assay Core Laboratory as described previously.5
Hematopoietic cell isolation
Fetal liver (FL) cells were isolated from embryos at E14.5. Limb tissue was genotyped by PCR for Gdf11 and Sry (to determine sex). Primers are listed in supplemental Table 1. Adult bone marrow (BM) cells were analyzed using a BD LSR II flow cytometer or BD FACS Aria II sorter (BD Biosciences). Antibodies, staining protocols, and gating strategies are detailed in supplemental Methods. Data were analyzed with FACS Diva v8.0.2 (BD) and FlowJo v10.1r5 software.
FL and BM transplantation
FL cells were pooled from 3 sex-matched embryos of the same genotype, and BM cells were pooled from 1 to 3 sex-matched donors of the same genotype. Total FL or BM cells (1 × 106) were injected retroorbitally into 8- to 10-week-old sex-matched lethally irradiated (9.5 Gy, split dose) CD45.1 (for FL cells) or C57BL/6J (for BM cells) congenic mice. Recipients were administered 0.67 mg/mL sulfamethoxazole and trimethoprim suspension in their drinking water for 4 weeks after injection.
Statistical analysis
Comparisons between 3 or more groups used 1-way analysis of variance (ANOVA) followed by Bonferroni posttest correction. Analyses over time used repeated measures ANOVA. Observed differences with P < .05 were considered statistically significant.
Results and discussion
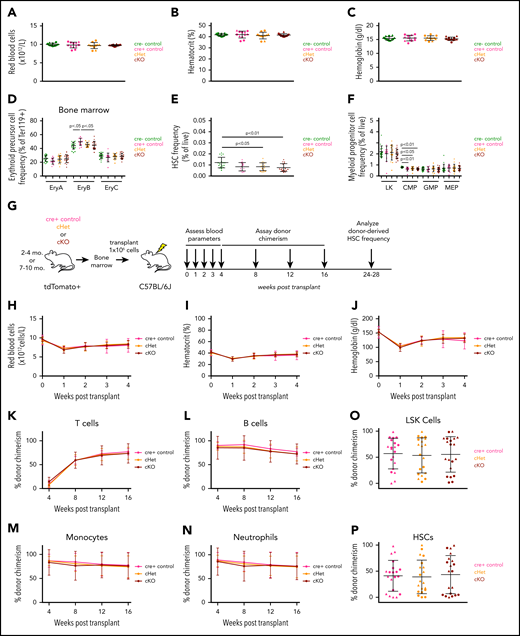
To investigate the role of GDF11 in normal hematopoiesis, we analyzed blood cell precursors in the FLs of Gdf11 knockout embryos (Gdf11−/−), which exhibit perinatal lethality.6 No differences were detected in overall cellularity (supplemental Figure 1a) or in the frequencies of erythroid precursor cell (EPC) subsets (Figure 1A-D) or hematopoietic stem cells (HSCs; Lin−Sca-1+c-Kit+CD48−CD150+; Figure 1E-H) among Gdf11−/−, Gdf11+/−, and Gdf11+/+ FLs, suggesting that hematopoietic development occurs normally in the absence of GDF11. We also assessed the effects of Gdf11 deletion in the context of hematopoietic stress, induced by FL HSPC transplantation (Figure 1I). The degree and kinetics of blood cell reconstitution, including recipient red blood cell (RBC) parameters (Figure 1J-L; supplemental Figure 1b-c) and peripheral leukocyte chimerism (Figure 1M-P), were similar among all donor genotypes. Body weight, spleen weight, and normalized heart weight were also equivalent among recipients (supplemental Figure 2a-f), whereas raw and normalized skeletal muscle weight was reduced in female, but not male, recipients of Gdf11−/− FL cells (supplemental Figure 2g-j). Assessment of recipient BM at 6 to 7 months posttransplant revealed similar frequencies of donor-derived LSK (Lineage−Sca-1+c-Kit+) progenitors and immunophenotypic HSCs, with all groups exhibiting a high rate of donor cell engraftment (average ∼90%) regardless of donor genotype (Figure 1Q-R). Thus, genetic disruption of Gdf11 during embryogenesis does not impact the ability of fetal HSPCs to repopulate the blood system after transplantation. In addition, although Gdf11 is normally expressed in a majority of adult peripheral blood cells, as well as minor subsets of BM and splenic precursors (supplemental Figure 3), adult animals engrafted with Gdf11−/− FL cells, in which the vast majority of blood lineage cells are unable to produce GDF11 protein, exhibit largely unperturbed hematopoietic (including erythropoietic) function and relatively normal appearance of nonhematopoietic organs. These mice also exhibited normal levels of circulating GDF11 and GDF8 protein (supplemental Figure 2k-l; see supplemental text), suggesting that systemic GDF11 derives largely from nonhematopoietic sources in this setting.
Whole-body Gdf11 deletion does not alter frequency of fetal hematopoietic stem and progenitor cells (HSPCs) or affect hematopoietic reconstitution following FL transplantation. (A-C) Representative CD71/Ter-119 flow cytometry plots of EPCs isolated from E14.5 (A) Gdf11+/+, (B) Gdf11+/−, and (C) Gdf11−/− FLs. Cells previously gated as live, Lin−, CD41−. EPCs were identified by differential expression of CD71 and Ter-119, with cells progressing in maturity from S1 to S5.20 (D) Quantification of EPC frequency among live cells in E14.5 Gdf11+/+, Gdf11+/−, and Gdf11−/− FLs (n = 12 embryos per genotype; males and females pooled). (E-G) Representative CD48/CD150 flow cytometry plots of HSCs isolated from E14.5 (E) Gdf11+/+, (F) Gdf11+/−, and (G) Gdf11−/− FLs. Cells previously gated as live, Lin−, Sca-1+, c-Kit+. (H) Quantification of HSC frequency among live cells in E14.5 Gdf11+/+, Gdf11+/−, and Gdf11−/− FLs (n = 12 embryos per genotype; males and females pooled). (I) Experimental design. Gdf11+/+, Gdf11+/−, and Gdf11−/− embryos were harvested at E14.5. FL cells from sex- and genotype-matched embryos were pooled, and 1 × 106 FL cells were injected IV into lethally irradiated sex-matched CD45.1 recipients. Recipients were monitored for blood parameters and donor chimerism levels as indicated. (J) RBC counts, (K) hematocrit, and (L) hemoglobin levels in recipient mice at 0 (baseline), 1, 2, 3, and 4 weeks postirradiation and transplant (n = 20 recipients per donor genotype; males and females pooled). (M-P) Monthly peripheral blood CD45.2+ donor chimerism levels within (M) CD3+ T cells, (N) B220+ B cells, (O) CD11b+ Ly6G− monocytes, and (P) CD11b+ Ly6G+ neutrophils (n = 20 recipients per donor genotype; males and females pooled). (Q-R) CD45.2+ donor cell chimerism levels within BM (Q) Lin−Sca-1+c-Kit+ cells and (R) Lin−Sca-1+c-Kit+CD48−CD150+ HSCs (n = 20 recipients per donor genotype; males and females pooled). Data are plotted as (J-P) mean ± standard deviation (SD) or as (D, H, Q-R) individual data points overlaid with mean ± SD. Statistics for panels J-P were calculated using a repeated-measures ANOVA. Statistics for panels D, H, Q-R were calculated using a 1-way ANOVA with Bonferroni posttest correction. For all panels, no differences with P < .05 were detected. (D, H, Q-R) Circles: males; triangles: females. LSK, Lineage−Sca-1+c-Kit+.
Whole-body Gdf11 deletion does not alter frequency of fetal hematopoietic stem and progenitor cells (HSPCs) or affect hematopoietic reconstitution following FL transplantation. (A-C) Representative CD71/Ter-119 flow cytometry plots of EPCs isolated from E14.5 (A) Gdf11+/+, (B) Gdf11+/−, and (C) Gdf11−/− FLs. Cells previously gated as live, Lin−, CD41−. EPCs were identified by differential expression of CD71 and Ter-119, with cells progressing in maturity from S1 to S5.20 (D) Quantification of EPC frequency among live cells in E14.5 Gdf11+/+, Gdf11+/−, and Gdf11−/− FLs (n = 12 embryos per genotype; males and females pooled). (E-G) Representative CD48/CD150 flow cytometry plots of HSCs isolated from E14.5 (E) Gdf11+/+, (F) Gdf11+/−, and (G) Gdf11−/− FLs. Cells previously gated as live, Lin−, Sca-1+, c-Kit+. (H) Quantification of HSC frequency among live cells in E14.5 Gdf11+/+, Gdf11+/−, and Gdf11−/− FLs (n = 12 embryos per genotype; males and females pooled). (I) Experimental design. Gdf11+/+, Gdf11+/−, and Gdf11−/− embryos were harvested at E14.5. FL cells from sex- and genotype-matched embryos were pooled, and 1 × 106 FL cells were injected IV into lethally irradiated sex-matched CD45.1 recipients. Recipients were monitored for blood parameters and donor chimerism levels as indicated. (J) RBC counts, (K) hematocrit, and (L) hemoglobin levels in recipient mice at 0 (baseline), 1, 2, 3, and 4 weeks postirradiation and transplant (n = 20 recipients per donor genotype; males and females pooled). (M-P) Monthly peripheral blood CD45.2+ donor chimerism levels within (M) CD3+ T cells, (N) B220+ B cells, (O) CD11b+ Ly6G− monocytes, and (P) CD11b+ Ly6G+ neutrophils (n = 20 recipients per donor genotype; males and females pooled). (Q-R) CD45.2+ donor cell chimerism levels within BM (Q) Lin−Sca-1+c-Kit+ cells and (R) Lin−Sca-1+c-Kit+CD48−CD150+ HSCs (n = 20 recipients per donor genotype; males and females pooled). Data are plotted as (J-P) mean ± standard deviation (SD) or as (D, H, Q-R) individual data points overlaid with mean ± SD. Statistics for panels J-P were calculated using a repeated-measures ANOVA. Statistics for panels D, H, Q-R were calculated using a 1-way ANOVA with Bonferroni posttest correction. For all panels, no differences with P < .05 were detected. (D, H, Q-R) Circles: males; triangles: females. LSK, Lineage−Sca-1+c-Kit+.
To further evaluate the functional impact of hematopoietic cell–derived Gdf11 expression on blood cell formation and maintenance, we produced conditional knockout mice (cKO) with pan-hematopoietic deletion of Gdf11 and tdTomato labeling of Cre-expressing cells using the vav-iCre,7 Gdf11fl/fl,8 and Rosa26-LSL-tdTomato Ai9 reporter alleles9 (supplemental Figure 4a-b). vav-iCre7 is expressed in the FL by E12.5 and is active postnatally in nearly all major hematopoietic lineages, including hematopoietic progenitors.10 vav-cKO (vav-iCre;Gdf11fl/fl;Rosa26tdTomato/+) and vav-conditional heterozygotes (cHet: vav-iCre;Gdf11fl/+;Rosa26tdTomato/+) were compared with 2 control genotypes (cre− control: Gdf11fl/fl;Rosa26tdTomato/+ and cre+ control: vav-iCre;Gdf11+/+;Rosa26tdTomato/+) (supplemental Figure 4c).
We validated our conditional deletion model by flow cytometry, genomic PCR, and real-time PCR, confirming a substantial reduction of splenic Gdf11 messenger RNA levels in cKO mice and a modest decrease in cHet mice (supplemental Figure 4d-j). Circulating GDF11 levels were also decreased ∼13% to 15% in cKO, but not cHet, mice, whereas GDF8 levels were unaltered across experimental groups (supplemental Figure 4k-l; see supplemental text). Also, although transcriptional adaptation has been reported for other loss-of-function alleles,11,12 we saw no signs that such genetic compensation occurs in Gdf11−/− or cKO mice (supplemental Figures 5 and 6).
No significant changes were noted in RBC parameters in cKO or cHet mice at 2 to 4 months or 7 to 10 months of age (Figure 2A-C; supplemental Figure 7). Frequencies of EPCs, Ter-119+ cells, and BM HSPCs were also unaffected in cKO or cHet mice relative to age-matched cre− and cre+ controls (Figure 2D-F; supplemental Figures 8 and 9; see supplemental text). vav–lineage-specific disruption of Gdf11 did not cause changes in body weight, BM cellularity, spleen weight, normalized heart weight, or raw or normalized muscle weight in mice at 2 to 4 months old, with most parameters similarly unperturbed in 7- to 10-month-old mice (supplemental Figure 10).
Hematopoietic cell–specific Gdf11 deletion does not alter adult erythropoiesis, RBC parameters, or HSPC frequency or hematopoiesis following adult BM transplantation. (A) RBC counts, (B) hematocrit, and (C) hemoglobin levels in 2- to 4-month-old mice (n = 10-12 mice per genotype; males and females pooled). (D-F) Frequency of BM (D) EPCs, (E) HSCs, and (F) myeloid progenitor cells within 2- to 4-month-old mice (n = 10-12 mice per genotype; males and females pooled). (G) Experimental design. BM was harvested from 2- to 4-month-old or 7- to 10-month-old Gdf11 cKO, cHet, and cre+ control mice. Cells from sex- and genotype-matched donors were pooled and 1 × 106 BM cells were injected IV into lethally irradiated sex-matched C57BL/6J recipients. Recipients were monitored for blood parameters and donor cell chimerism as indicated. (H) RBC counts, (I) hematocrit, and (J) hemoglobin levels within recipient mice at 0 (baseline), 1, 2, 3, and 4 weeks posttransplant (n = 16-20 recipients per donor genotype; males and females pooled). (K-N) tdTomato+ donor-derived peripheral blood cell chimerism levels determined monthly for (K) CD3+ T cells, (L) B220+ B cells, (M) CD11b+ Ly6G− monocytes, and (N) CD11b+ Ly6G+ neutrophils in transplant recipients (n = 16-20 recipients per donor genotype; males and females pooled). (O-P) tdTomato+ donor-derived BM cell chimerism levels for BM (O) Lin−Sca-1+c-Kit+ and (P) Lin−Sca-1+c-Kit+CD48−CD150+ cells in transplant recipients (n = 16-20 recipients per donor genotype; males and females pooled). Data are plotted as (H-N) mean ± SD or as (A-F, O-P) individual data points overlaid with mean ± SD. Statistics for panels H-N were calculated using a repeated-measures ANOVA. Statistics for panels A-F, O-P were calculated using a 1-way ANOVA with Bonferroni posttest correction. For all panels, no differences with P < .05 were detected. (A-F; O-P) Circles: males; triangles: females.
Hematopoietic cell–specific Gdf11 deletion does not alter adult erythropoiesis, RBC parameters, or HSPC frequency or hematopoiesis following adult BM transplantation. (A) RBC counts, (B) hematocrit, and (C) hemoglobin levels in 2- to 4-month-old mice (n = 10-12 mice per genotype; males and females pooled). (D-F) Frequency of BM (D) EPCs, (E) HSCs, and (F) myeloid progenitor cells within 2- to 4-month-old mice (n = 10-12 mice per genotype; males and females pooled). (G) Experimental design. BM was harvested from 2- to 4-month-old or 7- to 10-month-old Gdf11 cKO, cHet, and cre+ control mice. Cells from sex- and genotype-matched donors were pooled and 1 × 106 BM cells were injected IV into lethally irradiated sex-matched C57BL/6J recipients. Recipients were monitored for blood parameters and donor cell chimerism as indicated. (H) RBC counts, (I) hematocrit, and (J) hemoglobin levels within recipient mice at 0 (baseline), 1, 2, 3, and 4 weeks posttransplant (n = 16-20 recipients per donor genotype; males and females pooled). (K-N) tdTomato+ donor-derived peripheral blood cell chimerism levels determined monthly for (K) CD3+ T cells, (L) B220+ B cells, (M) CD11b+ Ly6G− monocytes, and (N) CD11b+ Ly6G+ neutrophils in transplant recipients (n = 16-20 recipients per donor genotype; males and females pooled). (O-P) tdTomato+ donor-derived BM cell chimerism levels for BM (O) Lin−Sca-1+c-Kit+ and (P) Lin−Sca-1+c-Kit+CD48−CD150+ cells in transplant recipients (n = 16-20 recipients per donor genotype; males and females pooled). Data are plotted as (H-N) mean ± SD or as (A-F, O-P) individual data points overlaid with mean ± SD. Statistics for panels H-N were calculated using a repeated-measures ANOVA. Statistics for panels A-F, O-P were calculated using a 1-way ANOVA with Bonferroni posttest correction. For all panels, no differences with P < .05 were detected. (A-F; O-P) Circles: males; triangles: females.
Finally, we evaluated the effects of vav-lineage Gdf11 deletion on hematopoietic repopulation following whole BM transplantation (Figure 2G). As with the FL transplants, we saw no differences in RBC recovery among recipients from the 2- to 4-month-old donor cohort (Figure 2H-J; supplemental Figure 11a-b). Most of these parameters were also unperturbed in recipients from 7- to 10-month-old donors (supplemental Figure 11c-g). Donor leukocyte and HSPC chimerism, assessed by tdTomato fluorescence, also showed no differences among experimental groups (Figure 2K-P). Multiple body parameters were indistinguishable among transplanted recipients (supplemental Figure 12), providing further support that Gdf11 disruption within the hematopoietic compartment does not induce overt pathology in the tissues analyzed.
In summary, our studies using multiple genetic mouse models suggest that GDF11 is largely dispensable for native and transplant-induced regenerative hematopoiesis. These findings are consistent with a recently published report13 applying vav-specific deletion of Gdf11 in the Hbbth3 model of β-thalassemia and in mice administered ActRII ligand traps. Our work argues against a prior model that EPC-derived GDF11 restrains erythropoiesis in homeostasis and in response to anemic stress1,2 and further contributes to ongoing investigations as to whether modulating GDF11 affects cardiac hypertrophy3,14-16 or muscle dysfunction.4,17-19 Furthermore, although blood-lineage cells may in some circumstances contribute to the circulating pool of GDF11 protein, loss of Gdf11 within hematopoietic lineage cells is generally insufficient to drive age-related cardiac or skeletal muscle phenotypes. Additional studies are needed to determine whether loss of Gdf11 from tissues outside of, or in addition to, the hematopoietic system may induce such phenotypes.
For original data, please contact amy_wagers@harvard.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S.-J. Lee for providing Gdf11fl/fl and Gdf11+/− mice, S. Bhasin and L. Peng for assistance with LC-MS/MS, M. J. Kim and L. D. Wang for technical assistance, and M. Tabebordbar, A. Almada, and G. Chen for helpful discussions.
This work was supported by National Institutes of Health (NIH), National Institute on Aging (NIA) grants R01AG048917 and R01AG057428 and a Glenn Foundation for Medical Research grant (A.J.W.), an NIH, NIA grant R01AG047131 (R.T.L.), an NIH Training grant T32 DK007529 (J.M.G.), an NIA Postdoctoral Fellowship grant F32 AG050395 (J.M.G.), and the John S. LaDue Memorial Fellowship in Cardiology (J.C.G.) and an NIH, National Heart, Lung, and Blood Institute Training grant T32 HL007572 (J.C.G.).
Authorship
Contribution: J.M.G. and A.J.W. designed the study; J.M.G, H.S., and K.A.M. performed most of the experiments, including flow cytometry and transplants, and analyzed data; M.F.-A. assisted with flow cytometry and real-time PCR analysis; J.C.G. and A.C.K. measured and analyzed cardiac weights; R.T.L. helped with experimental design; J.M.G. and A.J.W. interpreted data and wrote the manuscript; and all authors commented on the manuscript.
Conflict-of-interest disclosure: R.T.L. and A.J.W. are cofounders and members of the scientific advisory board and hold private equity in Elevian, a company that aims to develop medicines to restore regenerative capacity. Elevian provides sponsored research support to the Wagers and Lee labs. The remaining authors declare no competing financial interests.
Correspondence: Amy Wagers, Department of Stem Cell and Regenerative Biology, Harvard University and Harvard Stem Cell Institute, 7 Divinity Ave, Cambridge, MA 02138; e-mail: amy_wagers@harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal