In this issue of Blood, Ling et al have compared the function of 2 forms of the transcription factor GATA1 (the full-length protein and a shorter form). Their study shows that the N-terminal domain of GATA1 is critical to normal red cell differentiation (erythropoiesis).1
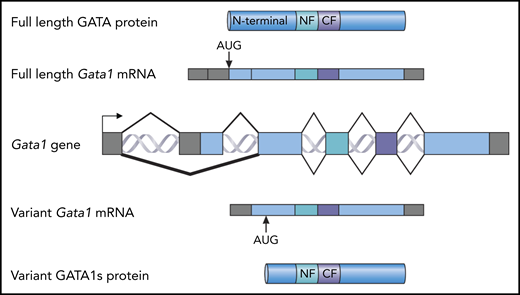
Full-length forms and short forms of the GATA1 protein. A map of the Gata1 locus is shown in the center (Gata1 gene). The exons are shown as blocks. Gray indicates noncoding sequence; blue indicates coding sequence. Above the gene, a spliced, full-length Gata1 messenger RNA (mRNA) is shown with the AUG initiation codon indicated. This RNA is translated into full-length GATA1 protein (top). The N-terminal domain, the N-terminal zinc finger (NF), and the C-terminal zinc finger (CF) are indicated. Below the gene, a spliced variant Gata1 mRNA encoding the variant GATA1s protein is shown, with the alternative GATA1s AUG initiation codon indicated. This RNA is translated as the GATA1s protein. The N-terminal zinc finger and the C-terminal zinc finger are indicated.
Full-length forms and short forms of the GATA1 protein. A map of the Gata1 locus is shown in the center (Gata1 gene). The exons are shown as blocks. Gray indicates noncoding sequence; blue indicates coding sequence. Above the gene, a spliced, full-length Gata1 messenger RNA (mRNA) is shown with the AUG initiation codon indicated. This RNA is translated into full-length GATA1 protein (top). The N-terminal domain, the N-terminal zinc finger (NF), and the C-terminal zinc finger (CF) are indicated. Below the gene, a spliced variant Gata1 mRNA encoding the variant GATA1s protein is shown, with the alternative GATA1s AUG initiation codon indicated. This RNA is translated as the GATA1s protein. The N-terminal zinc finger and the C-terminal zinc finger are indicated.
For those who study erythropoiesis, the foundational role of GATA1 is well known.2 Originally, the GATA1 protein was identified solely by its ability to bind DNA. After the gene encoding the protein was cloned, it was quickly shown that deletion of the Gata1 gene in transgenic mice caused a lethal anemia, resulting in death at the fetal liver stage.3 GATA1 is a member of a family of zinc finger proteins that share a similar structure. The C-terminal zinc finger is primarily responsible for the binding of GATA1 to the sequence WGATAR. The N-terminal finger stabilizes the binding to DNA and also facilitates the interaction with FOG1, which is 1 of several proteins that interact with GATA1. The N-terminal domain of GATA1 interacts with other proteins, including RUNX1 and RB4 (see figure).
Mutations in GATA1 are associated with many hematologic diseases.5 In particular, mutations in the second exon lead to an alternative splicing event or altered translation that essentially deletes the N-terminal domain of GATA1, which results in a shorter protein, GATA1 short (GATA1s). Germline mutations in exon 2 result in a form of Diamond-Blackfan anemia,6 and somatic mutations are seen in Down syndrome acute megakaryocytic leukemia.5
These observations lead to the question: What is the functional role of GATA1s? To address this question, Ling et al used a transgenic mouse that was engineered to exclusively express GATA1s.7,8 These mice show an inhibition of erythropoiesis and a transient expansion of megakaryocyte progenitors at the fetal liver stage of development, similar to what is seen in Down syndrome patients. Ling et al performed multi-omics profiling of GATA1s and wild-type fetal liver cells. They found that although the transcriptional profiles, the GATA1 occupancy, histone modification, and chromatin accessibility profiles were largely overlapping between the GATA1s and wild-type fetal liver cells, the loss of the N terminus of GATA1 specifically altered the expression of a subset of genes, which was accompanied by changes in GATA1s occupancy and/or chromatin modifications. These data clearly demonstrate a specific role for the N- terminal domain of GATA1 in normal erythropoiesis. Ling et al provide a number of targets and pathways that will be important to investigate in the future.
In normal hematopoiesis, GATA2 occupies WGATAR sites in the genome in primitive cells before being replaced by GATA1 as cells differentiate down the erythroid lineage.2 GATA1s mice expressed higher levels of Gata2 and Runx1 messenger RNA, leading to the hypothesis that the lingering expression of GATA2 was inhibiting erythropoiesis by preventing the binding of GATA1s. To test this, Ling et al crossed their Gata1s mice to mice carrying a Gata2 knockout allele. They found that erythropoiesis was rescued in Gata1s mice carrying a Gata2 null allele, demonstrating that Gata1-mutant embryos can be rescued by haploinsufficiency for GATA2.
The Ling et al study is an example of both hypothesis-testing (role of the GATA1 N-terminal domain) and hypothesis-generating (identification of GATA1 N-terminal domain regulated pathways) results. It will be important to determine the features of the N-terminal domain of GATA1 that are required for normal erythropoiesis. Identifying the proteins that interact with the N-terminal domain of GATA1 will be critical to our understanding of how GATA1 mutations that result in the deletion of the N-terminal domain cause hematologic diseases. Ling et al have made an impressive start in answering this question.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal