Because hematopoietic stem cells (HSCs) were found to require a bone marrow (BM) habitat for long-term function, many studies have attempted to dissect key cellular and molecular interactions between HSCs and their BM microenvironments or HSC niches. In this issue of Blood, Mende et al provide a computational method to infer potential ligand-receptor interactions between murine hematopoietic stem and progenitor cells (HSPCs) and their niche-forming cells.1
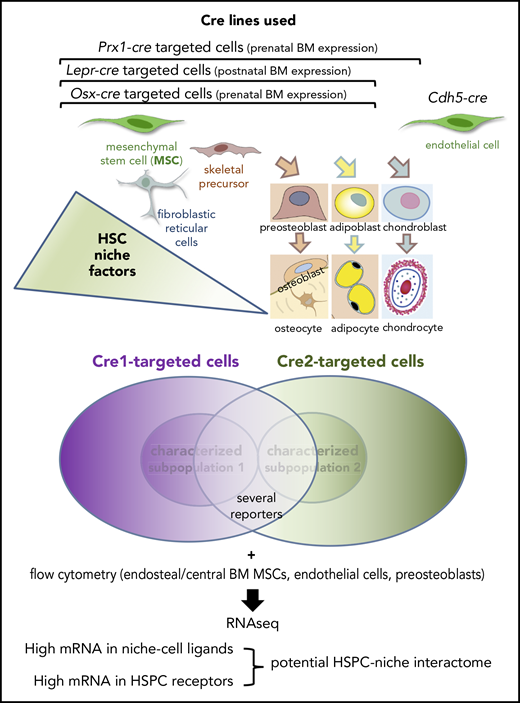
Mende et al use a variety of Cre lines targeting multiple nonhematopoietic cells of the BM microenvironment. They attempt to resolve previous discrepancies in the field whereby the overlap between niche cells targeted by different Cre lines was underestimated because only subsets among those Cre-targeted cells had been characterized. To achieve their goal, they used several reporters and performed RNA sequencing from immunophenotypically identified HSPCs, mesenchymal stromal cells, endothelial cells, and preosteoblasts isolated from bone-associated (endosteal) BM or located further from bone (central). Matching top expressed ligands in niche cells with the most highly expressed receptors on HSPCs provides a candidate HSPC-niche interactome containing some old and many new potential HSPC regulators.
Mende et al use a variety of Cre lines targeting multiple nonhematopoietic cells of the BM microenvironment. They attempt to resolve previous discrepancies in the field whereby the overlap between niche cells targeted by different Cre lines was underestimated because only subsets among those Cre-targeted cells had been characterized. To achieve their goal, they used several reporters and performed RNA sequencing from immunophenotypically identified HSPCs, mesenchymal stromal cells, endothelial cells, and preosteoblasts isolated from bone-associated (endosteal) BM or located further from bone (central). Matching top expressed ligands in niche cells with the most highly expressed receptors on HSPCs provides a candidate HSPC-niche interactome containing some old and many new potential HSPC regulators.
Niches formed by mesenchymal stem cells (MSCs) associated with endothelial cells were suggested a decade ago as critical regulators of HSCs.2,3 In the mouse, different genetic models have been used to mark MSCs, and it is agreed that these cells are an important source of key HSC factors.4,5 However, a systematic comparison among HSC niche cells that were identified by using different strategies has been lacking until recently. Moreover, single-cell technologies have subsequently revealed that MSCs do not escape the large heterogeneity observed in other stem cell populations.6,7 As a consequence, some discrepancies remain regarding the actual source and distribution of prominent HSC factors. The variable specificity, expression, and penetrance across genetic models is a cause for debate. For example, combined deletion of essential HSC factors in multiple BM cell types (or the bulk of nonhematopoietic BM stroma) using constitutive Cre lines reduces HSCs4,5 but cannot be compared with much smaller deletions in 1-cell populations targeted (among other cells) by those Cre lines, and this approach has limited power to uncover specific HSC niche-cell interactions. The diversity of HSC niches is partly anatomical because different types of blood vessels and associated perivascular cells have been found in different BM regions, such as those close to the bone surface (endosteal) and those further away from the bone (central). However, whether stromal or endothelial cells in different BM regions interact distinctly with HSCs has not been addressed in a systematic way.
Mende et al undertake the impressive task of cross-comparing mesenchymal stromal cells and endothelial cells defined immunophenotypically and using genetic drivers in cell populations isolated from the central and the endosteal BM of mice. Through their systematic assessment they confirm the overlap of HSC niche-cell populations identified by using different strategies (which had been suggested in previous studies) and identify interesting differences among cell populations harvested from endosteal and central BM (see figure). For instance, MSCs harvested from central or endosteal BM show distinct transcriptional profiles. Together with the reportedly higher resistance of endosteal MSCs to myeloablation and the described functions of some endosteal mesenchymal cells such as N-cadherin+ cells,8 these results add further evidence to the possibility that different BM niches might regulate steady-state vs stress hematopoiesis.
As a major novel aspect, the authors develop a computational method to infer potential ligand-receptor interactions between niche cells and HSPCs based on messenger RNA (mRNA) expression levels. A ranking and matching algorithm assigns highly expressed ligands (either agonists or antagonists) in niche cells to putative receptors in HSPCs. Confirmation of key known interactions validates this method, which suggests potential novel interactions between HSPCs and their niches.
Previous studies have cross-compared the gene expression signatures of murine HSC-supporting and -nonsupporting stromal cell lines and have identified a modular network of paracrine signaling that includes most known and potential new HSC regulators, which have been subsequently tested.9 After identifying new candidate interactive partners between HSCs and some niche cells, it would be interesting to functionally test them. The Wascow laboratory has already started this task by investigating the effect of several regulators on HSPCs in vitro.
The study by Mende et al suggests differential interactions between HSPCs and CD31hiCD144+ (putative arteriolar) endothelial cells or MSCs, so it would be interesting to expand these analyses to other HSC niche cells, such as CD31lo sinusoidal endothelial cells and megakaryocytes. It is worth mentioning that this extensive cross-comparison of mouse models and prospectively identified HSC niche cells has been performed at the cell population level. After the development of HSC niche single-cell technologies,6,7 a natural extension of the Mende et al study would be applying these interactome algorithms to single-cell transcriptomics and/or examining candidate interactome pathways using additional markers and single-cell RNA sequencing (or index sorting) to discriminate the specific cell types critically interacting with HSCs through novel pathways.
On a practical note, it would be very useful for the research community to develop a publicly available online resource to facilitate the application of the elegant mathematical algorithm developed by Mende et al to the analyses of ligand-receptor interactions in other data sets from different laboratories. In that regard, other publicly available resources, such as the HSC niche database (http://stemniche.snv.jussieu.fr/), which allows interrogation of mRNA or microRNA expression in murine HSC-supporting stromal cells,9 or an interactive browser to interrogate gene expression in single BM vascular, perivascular, and osteolineage cells (http://aifantislab.com/niche/)6 have proven useful for investigating HSC niche-cell transcriptomics.
Future efforts in this area will need to include proteomics analysis because mRNA expression does not always correlate with protein abundance. This is particularly relevant for cytokine or chemokine receptors, which are often regulated by posttranscriptional mechanisms. Therefore, an important challenge for future studies is the extrapolation of candidate receptor-ligand interactions inferred from mouse transcriptomics to the human proteome. In this regard, age-associated alterations of HSPCs and their niches described in the human proteome resemble those previously found through mouse genetics,10 suggesting the potential translatability of these findings in general.
Conflict-of-interest disclosure: S.M.-F. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal