The World Health Organization (WHO) diagnostic criteria for chronic myelomonocytic leukemia (CMML) include clinical and morphological features; however, demonstrating clonality is not an absolute requirement for making the diagnosis.1 In this issue of Blood, Cargo et al show that patients with clonal monocytosis identified by targeted gene sequencing have a clinical outcome similar to that of overt WHO-defined CMML.2
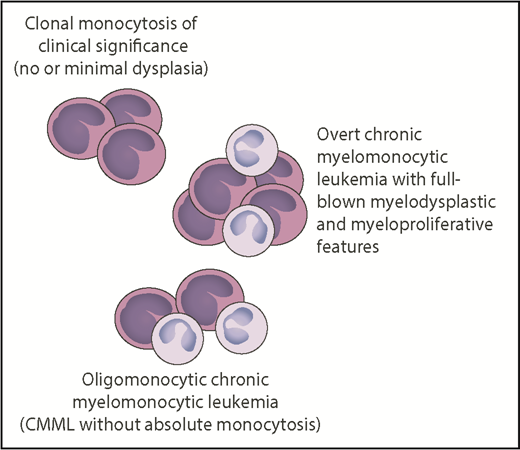
Relationship between clonal monocytosis of clinical significance, oligomonocytic CMML, and overt CMML. The number of monocytes reflects monocytic proliferation, whereas myelodysplasia is represented by neutrophils with hypogranulated cytoplasm and bilobed nucleus. Somatic mutations in genes like TET2, SRSF2, ASXL1, NRAS, KRAS, CBL, or SETBP1 represent the common thread of these chronic myeloid neoplasms, whereas epigenetic factors may be responsible for the phenotypic variability. Professional illustration by Patrick Lane, ScEYEnce Studios.
Relationship between clonal monocytosis of clinical significance, oligomonocytic CMML, and overt CMML. The number of monocytes reflects monocytic proliferation, whereas myelodysplasia is represented by neutrophils with hypogranulated cytoplasm and bilobed nucleus. Somatic mutations in genes like TET2, SRSF2, ASXL1, NRAS, KRAS, CBL, or SETBP1 represent the common thread of these chronic myeloid neoplasms, whereas epigenetic factors may be responsible for the phenotypic variability. Professional illustration by Patrick Lane, ScEYEnce Studios.
In the WHO criteria, CMML is classified as a myelodysplastic/myeloproliferative neoplasm (MDS/MPN), a category that also includes atypical chronic myeloid leukemia, juvenile myelomonocytic leukemia, and the MDS/MPN with ring sideroblasts and thrombocytosis.1 These disorders have both myelodysplastic (dysplasia and cytopenia) and myeloproliferative features (“cytosis” of 1 or more myeloid lineages) at the time of diagnosis.
CMML is characterized by the accumulation of monocytes in the peripheral blood, and therefore, the initial diagnostic approach involves the differential diagnosis of monocytosis. Once reactive monocytosis has been excluded, the possibility of CMML should be considered, especially if the elevated monocyte count has persisted for ≥3 months. According to the WHO criteria, diagnosis of CMML requires an absolute monocyte count ≥1 × 109/L with monocytes accounting for ≥10% of circulating leukocytes. These cutoffs are arbitrary: the natural history of disease is that the monocyte count increases from normal to elevated in a continuous manner. Monocytosis can be present in other myeloid malignancies, such as MPNs, and therefore, diagnosis of CMML requires the exclusion of these conditions. To establish the myelodysplastic nature of the disease, the presence of dysplasia involving ≥1 myeloid lineages is required, whereas blasts must constitute <20% of the cells in the peripheral blood and bone marrow.
As CMML lacks a unique disease-defining genetic lesion, genetic data have so far played a minor role in the diagnosis.1 About three-quarters of patients have a normal karyotype, which means that cytogenetic abnormalities can be used as clonal markers only in a subset of patients.3 Somatic gene mutations have been identified only in the last few years. A recent study using a panel of 38 recurrently mutated genes in myeloid malignancies has detected somatic mutations in 199 of 214 CMML patients (93%).4 The most frequently mutated genes were TET2, SRSF2, ASXL1, NRAS, KRAS, and SETBP1. A significant association was found between mutations in TET2 and spliceosome genes, and one-fifth of patients showed cooccurrence of TET2 and SRSF2 mutations, a comutation pattern that can be considered relatively typical of CMML. Quantification of monocyte subsets by flow cytometry has recently provided a new tool for the diagnosis of CMML.5 An increase in the fraction of classical monocytes (CD14++/CD16−) to >94.0% of total monocytes has been found to be a biomarker that helps distinguish CMML from reactive monocytosis.
Cargo et al conducted a study that generated from routine hematology practice. They studied samples of patients referred to a hematology service for monocytosis. Through targeted sequencing of 27 genes recurrently mutated in myeloid malignancies, they detected ≥1 somatic mutation in 221 of 283 samples (78%). Overall, 207 subjects underwent additional tests, including bone marrow assessment, for a definitive diagnosis. Virtually all patients with a confirmed myeloid neoplasm carried a somatic mutation (140/142; 99% of cases), and most of them had CMML (114/142; 80% of cases). Of the 65 subjects who did not have a definitive diagnosis but just indeterminate features, 37 (57% of cases) carried at least 1 somatic mutation, with TET2, SRSF2, and ASXL1 being the most frequently mutated genes. In terms of variant allele frequency (VAF), there was no significant difference between the diagnostic and nondiagnostic/indeterminate features groups, with average values ∼40%. More importantly, the overall survival of mutated nondiagnostic patients was indistinguishable from that of patients with WHO-defined CMML and worse than that of subjects with monocytosis without somatic mutations. Flow cytometry analysis of circulating monocytes showed overlapping features in mutated nondiagnostic subjects and CMML patients.
The study by Cargo et al validates the current WHO diagnostic criteria for CMML, showing that when myelodysplasia is absent or minimal, the diagnosis of CMML may still be made if a somatic mutation is present. In fact, many of the cases that were initially considered nondiagnostic based on morphological criteria were identified by demonstrating an acquired clonal genetic abnormality, as stipulated in the recently revised WHO criteria.1 The conclusions of the study, however, go beyond this validation and suggest that the presence of a somatic mutation should become an absolute requirement for diagnosis of CMML, irrespective of the presence or absence of dysplasia. The fact that somatic mutations of myeloid genes can be found also in healthy individuals with age-related clonal hematopoiesis (ARCH)6 does not represent a valid reason for not using them as markers of clonality in myeloid neoplasms. In both patients with clonal monocytosis and those with CMML, the VAF of somatic mutations was much higher (∼40% on average) than that commonly observed in healthy subjects with ARCH (<10%), indicating a much more advanced clonal disease.
Through their investigations, Cargo et al have illuminated a condition that can be defined as “clonal monocytosis of clinical significance.” The relationship between this condition and CMML resembles that between clonal cytopenia of undetermined significance (CCUS) and MDS.7,8 The overall survival and the risk of disease progression of patients with CCUS and highly specific mutation patterns are indistinguishable from those of patients with a myeloid neoplasm with myelodysplasia.9 While clonal monocytosis of clinical significance lacks overt myelodysplasia, an oligomonocytic CMML has also been described that displays a similar clinicopathologic and mutational profile to classical CMML.10 Somatic mutations represent the common thread of all these conditions, which are schematically represented (see figure).
In conclusion, the available evidence suggests that demonstrating somatic mutations and defining their patterns may provide presumptive evidence of myeloid malignancies, specifically, of CMML, even in the absence of definitive morphological criteria. In addition, integrating clinical features, morphology, immunophenotyping, and gene mutations may also improve risk stratification of these patients, providing a robust basis for clinical decision making and a reliable tool for clinical trials.4
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal