Key Points
Newly diagnosed primary cHLs have a concomitant increase in CD4+ Th1-polarized Tregs and differentiated Teffs.
Primary cHLs exhibit 2 major complementary bases of immunosuppression: likely exhausted PD-1+ Th1 Teffs and active PD-1− Th1 Tregs.
Abstract
In classical Hodgkin lymphoma (cHL), the host antitumor immune response is ineffective. Hodgkin Reed-Sternberg (HRS) cells have multifaceted mechanisms to evade the immune system, including 9p24.1/CD274(PD-L1)/PDCD1LG2(PD-L2) genetic alterations, overexpression of PD-1 ligands, and associated T-cell exhaustion and additional structural bases of aberrant antigen presentation. The clinical success of PD-1 blockade in cHL suggests that the tumor microenvironment (TME) contains reversibly exhausted T effector cells (Teffs). However, durable responses are observed in patients with β2-microglobulin/major histocompatibility complex (MHC) class I loss on HRS cells, raising the possibility of non-CD8+ T cell–mediated mechanisms of efficacy of PD-1 blockade. These observations highlight the need for a detailed analysis of the cHL TME. Using a customized time-of-flight mass cytometry panel, we simultaneously assessed cell suspensions from diagnostic cHL biopsies and control reactive lymph node/tonsil (RLNT) samples. Precise phenotyping of immune cell subsets revealed salient differences between cHLs and RLNTs. The TME in cHL is CD4+ T-cell rich, with frequent loss of MHC class I expression on HRS cells. In cHLs, we found concomitant expansion of T helper 1 (Th1)-polarized Teffs and regulatory T cells (Tregs). The cHL Th1 Tregs expressed little or no PD-1, whereas the Th1 Teffs were PD-1+. The differential PD-1 expression and likely functional Th1-polarized CD4+ Tregs and exhausted Teffs may represent complementary mechanisms of immunosuppression in cHL.
Introduction
Classical Hodgkin lymphomas (cHLs) consist of rare malignant Hodgkin Reed-Sternberg (HRS) cells embedded within an extensive inflammatory/immune cell infiltrate. Despite the paucity of HRS cells and the brisk immune cell infiltrate, there is little evidence of an effective antitumor immune response in cHL.
HRS cells evade antitumor immunity by multiple mechanisms, including copy gain of chromosome 9p24.1/CD274(PD-L1)/PDCD1LG2(PD-L2) and copy number–dependent increased expression of the PD-1 ligands.1-3 Recent clinical trials revealed the sensitivity of cHL to PD-1 blockade.4-7 However, the mechanism of action of PD-1 blockade in this disease remains to be defined.
In certain human solid tumors and additional murine models, the efficacy of PD-1 blockade has been linked to CD8+ cytotoxic T-cell activation in the tumor microenvironment (TME).3,8-11 CD8+ cytotoxic T cells recognize tumor antigens presented by major histocompatibility complex (MHC) class I molecules that are transported to the cell surface in association with β2-microglobulin (β2M). However, HRS cells frequently exhibit copy loss or inactivating mutations of B2M.12 Consistent with these findings, HRS cells often have absent or decreased cell surface expression of β2M and MHC class I,3,12,13 calling into question the importance of CD8+ effector cells for the activity of PD-1 blockade. In contrast, recent studies highlight the possible role of MHC class II–mediated antigen presentation to CD4+ effector cells in antitumor immunity.14-17 HRS cells frequently retain MHC class II expression,3 likely as a result of their derivation from MHC class II+ germinal center B cells.18 Additionally, in the intact cHL microenvironment, PD-L1+ HRS cells are significantly more likely to be in physical proximity to PD-1+ CD4+ T cells than to PD-1+ CD8+ T cells.19
Consistent with these observations, we recently found that HRS cell expression of β2M and MHC class I was not predictive for complete remission or progression-free survival in patients with relapsed/refractory cHL who were treated with PD-1 blockade (nivolumab).3 However, HRS cell expression of MHC class II was predictive for complete remission and prolonged progression-free survival following PD-1 blockade in patients with fully reconstituted immune systems,3 highlighting the potential role of CD4+ T cells in the cHL TME. For these reasons, we have performed a detailed analysis of CD4+ T cells and the inflammatory/immune cell infiltrate in primary cHLs using a customized time-of-flight mass cytometry (CyTOF) panel.
Materials and methods
Tissue samples
Lymph node biopsies from 7 patients with newly diagnosed cHL and lymph node or tonsil specimens from 10 patients with reactive lymphoid hyperplasia, but no evidence of malignant disease, were collected at the University of Washington. Pathology reports and Epstein-Barr virus (EBV) status of the cHL cases are summarized in supplemental Table 1, available on the Blood Web site. Institutional review board approval was obtained for analysis of these patient-derived samples. Viable lymph node or tonsil suspensions were prepared and cryopreserved as previously described.20
Antibodies
Mass cytometry antibodies and reporter isotopes are included in supplemental Table 2 and are described in detail in supplemental Methods.
CyTOF sample preparation
Separate cell surface and intracellular antibody master solutions were freshly prepared for each CyTOF run. Every run included a technical control of a peripheral blood mononuclear cell (75%) and cHL cell line, KMH2, (25%) admixture.
Each primary cHL or reactive lymph node/tonsil (RLNT) sample was partially thawed at 37°C rapidly and resuspended in warmed RPMI 1640 supplemented with fetal bovine serum (1:1 volume/volume). Cells were centrifuged twice for 10 minutes at 300g and passed through a 50-μm filter between centrifugation steps. The cell pellet was resuspended in 1 mL of RPMI 1640. Cells were stained for viability with 5 mM cisplatin for 2 minutes at room temperature and quenched with RPMI 1640 supplemented with 10% fetal bovine serum (5:1 volume/volume).
Cells were washed once with cell-staining media (CSM; 500 mL of barium-free phosphate-buffered saline [Gibco], 2.5 g of bovine serum albumin [Sigma], 100 mg of sodium azide [Sigma], 2 mL of 500 μM EDTA [Gibco]) and incubated for 10 minutes at room temperature with human FcR Blocking Reagent (Miltenyi Biotec). Cells were stained with the surface antibody cocktail (supplemental Table 2) for 30 minutes and washed once with CSM. Thereafter, cells were permeabilized with FoxP3 Fix/Perm Buffer (eBioscience) by gently shaking at room temperature in the dark. Cells were washed twice with eBioscience Wash Buffer (800g for 5 minutes), incubated with the intracellular antibody cocktail (supplemental Table 2) for 45 minutes at room temperature, and washed again. Thereafter, cells were incubated overnight at 4°C in 1 mL of 1:2000 Cell-ID Intercalator-Ir (Fluidigm) diluted in phosphate-buffered saline with 1.6% paraformaldehyde and 0.3% saponin (Sigma).
Cells were then washed twice with CSM, once with water, and resuspended at a concentration of 1 million cells per mL in deionized water containing a 1:10 dilution of EQ Four Element Calibration Beads (Fluidigm). Cells were filtered through a 35-μm membrane prior to mass cytometry acquisition.
Mass cytometry data analysis
CyTOF data acquisition is described in detail in supplemental Methods. The data were analyzed using the X-shift clustering algorithm, which was run as part of the VorteX clustering and visualization environment (version VorteX 29-Jun-2017-rev2).21 The number of events to be sampled was set by the maximum available cell numbers in the smallest sample to avoid skewing the data toward larger samples. These events were then combined into a single file prior to clustering to enable comparison between samples.
The Cytobank platform was used to first manually gate and identify the relevant populations to export. Two separate X-shift analyses were performed: all viable singlet cells, sampling 15 000 events, and CD3+ cells, sampling 7350 events. The CD3+ population was downsampled to ensure that equal numbers of events were captured from all cases. All antibody channels were used to perform the clustering in the viable singlet population. For the CD3+ cell analysis, all antibody channels with the exception of PAX5, CD163, CD14, and CD68 were used.
X-shift cluster visualization
The identified clusters were visualized by randomly sampling a proportional number of events from each cluster and generating force-directed layouts (FDLs). Each cluster was labeled with a unique color based on hex color code generator software (http://www.color-hex.com).
Heat maps were used to visualize the protein expression profiles of the identified clusters. For a given cluster, we chose the median expression level across all sampled cells to be the cluster protein expression value. These values were collated to form an overall protein expression matrix and normalized into z-scores, ranging from −4 to +4, using the “scale” function in R. Biclustering implemented in the “pheatmap” package then ordered similar columns and rows together in the expression matrix, according to the Pearson correlations for initial clustering distances and the complete distance for updated distances by the “hclust” function.
Using the heat maps and raw abundance data from X-shift, we phenotypically labeled each cluster according to well-defined lineage, differentiation, and polarization markers. For clusters with shared phenotypes, additional markers were used to further define differences. For inclusion in downstream analysis, we applied a cutoff ≥ 5% of sampled events in each cluster. Clusters without a clearly defined subtype that met this criterion were classified as “other.” Every sample in the analysis contributes a varying number of cells to each cluster. To compare differences between cHLs and RLNTs, we grouped them separately and took the median of each group.
Manual analysis of HRS cells
Using the Cytobank platform, HRS cells were identified and gated within the nonsinglet cluster. Histograms were generated to examine expression of phenotypic markers of HRS cells.
The CD15+ CD30+ population was exported to Excel for analysis of individual cells. Events were sorted into those that coexpressed CD3+ and those that had no expression of CD3+. These 2 groups were analyzed for expression of β2M and MHC class I, and the raw signal intensity of every HRS cell was plotted using GraphPad Prism.
Statistical methods
The distribution of cell counts between RLNT samples and cHL specimens was assessed using a Wilcoxon rank-sum test. Nominal P values were reported for exploratory or supplemental analyses; P values < .05 were considered statistically significant. Statistical analyses were performed using R (version 3.3.2).
Mass cytometry antibodies, peripheral blood mononuclear cell isolation, in vitro activation of normal T cells, macrophage differentiation and polarization, cell lines, CyTOF data acquisition, EBV-encoded small RNA in situ hybridization, EBV polymerase chain reaction analysis, and immunohistochemistry are described in supplemental Methods.
Results
CyTOF panel for simultaneous assessment of HRS cells and the TME
We assembled a panel of 39 isotope-conjugated antibodies to characterize the HRS cells and the associated inflammatory/immune cell infiltrate at the single-cell level. Our panel includes validated commercial reagents that assess lineage, differentiation, and polarization of certain T-cell subsets. To this, we added antibodies to identify other T-cell subtypes and capture additional functional attributes of cytotoxicity, activation, and exhaustion (Figure 1; supplemental Table 2). We also included reagents to characterize HRS cells (CD15, CD30, and PAX5) and evaluate their antigen presentation pathway components (β2M, pan–MHC class I, pan–MHC class II), PD-1 ligand expression, and JAK/STAT activity (phosphorylated STAT1 [pSTAT1]) (Figure 1). Lastly, we added antibodies to identify other cellular components of the immune system, such as B cells, macrophages, and natural killer (NK) cells (Figure 1).
CyTOF panel for the simultaneous assessment of HRS cells and infiltrating immune cells. The CyTOF panel identifies cells of T, B, NK, and macrophage lineages. CD4+ and CD8+ T cells can be further discriminated based on differentiation, polarization, and functional status. Malignant HRS cells are defined by their expression of CD30+ and CD15+, in addition to PAX5, PD-L1, PD-L2, and pSTAT1.
CyTOF panel for the simultaneous assessment of HRS cells and infiltrating immune cells. The CyTOF panel identifies cells of T, B, NK, and macrophage lineages. CD4+ and CD8+ T cells can be further discriminated based on differentiation, polarization, and functional status. Malignant HRS cells are defined by their expression of CD30+ and CD15+, in addition to PAX5, PD-L1, PD-L2, and pSTAT1.
The final panel (Figure 1; supplemental Table 2) was confirmed to function as predicted in CyTOF analyses of an admixture of the cHL cell line, KMH2, and normal donor peripheral blood mononuclear cells. Nucleated cells were identified with natural 191Iridium and 193Iridium (Ir191/193), a DNA cationic intercalator that binds to cellular nucleic acid. Multinucleated HRS cells have higher Ir191/193 uptake and largely reside in the nonsinglet cell fraction (supplemental Figure 1). HRS cells are often encircled by a “rosette” of T cells, an additional reason why HRS cells reside within the nonsinglet fraction (supplemental Figure 1).
CyTOF analyses of primary HRS cells
Given the known scarcity of HRS cells in viable tumor suspensions, the presumptive HRS cell population was analyzed manually. We identified viable HRS cells in the nonsinglet fraction of each primary cHL cell suspension based on HRS cell coexpression of CD30+ and CD15+ (Figure 2). The CD30+/CD15+ HRS cell populations were rare, representing <1% of sampled events. As expected, manually gated CD4+ T cells, CD8+ T cells, and PAX5+ B cells (from the singlet fraction) lacked CD30 and CD15 expression (Figure 2B). The identified CD30+/CD15+ HRS cells also expressed PAX5, PD-L1, and pSTAT1, characteristic features of malignant HRS cells1,2,22 (Figure 2B).
Identification and characterization of HRS cells. (A) A representative cHL with CD15+/CD30+ HRS cells identified by manual gating. (B) HRS cells from each analyzed primary cHL (1-7). HRS cells express CD15+ and CD30+. CD4+ T cells, CD8+ T cells, or B cells from the same primary cHL samples lack CD15+ and CD30+ expression. PAX5+ is expressed by HRS cells and B cells. PD-L1 and pSTAT1 are expressed by HRS cells from all examined cHLs. These markers are absent or reduced in the other cell subsets.
Identification and characterization of HRS cells. (A) A representative cHL with CD15+/CD30+ HRS cells identified by manual gating. (B) HRS cells from each analyzed primary cHL (1-7). HRS cells express CD15+ and CD30+. CD4+ T cells, CD8+ T cells, or B cells from the same primary cHL samples lack CD15+ and CD30+ expression. PAX5+ is expressed by HRS cells and B cells. PD-L1 and pSTAT1 are expressed by HRS cells from all examined cHLs. These markers are absent or reduced in the other cell subsets.
Aberrant expression of MHC class I on HRS cells detected by CyTOF
To interrogate MHC class I expression on HRS cells, we took advantage of the fact that our cHL cell suspensions included “bare” HRS cells and HRS cells encircled by adherent T cells (T-cell rosettes). We used a gating strategy similar to that described in previous cHL flow cytometric analyses.20,23 First, we gated the nonsinglet cells on the basis of CD45 and CD3 expression (Figure 3A, left panel). After confirming that the CD3− cells also lacked CD4 and CD8 expression, we positively gated on the CD3− subset that coexpressed CD30 and CD15, the “bare” HRS cells (Figure 3A, middle and lower right panels). The CD45+/CD3+ fraction also contained a subset of cells that coexpressed CD30 and CD15, rosetted HRS cells (Figure 3A, upper right panel). Thereafter, we confirmed that the positively selected CD30+/CD15+ HRS cell/T-cell rosettes were also CD3+, whereas the bare HRS cells were CD3− (Figure 3B).
Analysis of β2M and MHC class I expression on HRS cells by CyTOF. (A) Rosetted HRS cells separated from bare HRS cells by sequential gates. Events from the nonsinglet population are split according to CD3+ expression. Events within the CD3− gate are further gated to identify the CD4−/CD8− population. The CD3−/CD4−/CD8− subset was positively gated to select cells that coexpressed CD30 and CD15, representing bare HRS cells. Within the CD3+ population, CD15+/CD30+ coexpression identifies rosetted HRS cells. (B) CD3+ expression on rosetted HRS cells (upper panels) and bare HRS cells (lower panels). β2M (C) and MHC class I (D) expression on bare HRS cells (cyan) and CD3+ rosetted HRS cells (dark blue) in each of the primary cHLs (1-7). Significant differences were identified using the Wilcoxon rank-sum test. (E) Dual immunohistochemical analysis (PAX5 [red] and MHC class I [brown]) of primary cHLs (cases 2, 5, and 7) and a cHL with known HRS cell expression of MHC class I (positive control [Pos.]); arrows denote HRS cell membranes.3
Analysis of β2M and MHC class I expression on HRS cells by CyTOF. (A) Rosetted HRS cells separated from bare HRS cells by sequential gates. Events from the nonsinglet population are split according to CD3+ expression. Events within the CD3− gate are further gated to identify the CD4−/CD8− population. The CD3−/CD4−/CD8− subset was positively gated to select cells that coexpressed CD30 and CD15, representing bare HRS cells. Within the CD3+ population, CD15+/CD30+ coexpression identifies rosetted HRS cells. (B) CD3+ expression on rosetted HRS cells (upper panels) and bare HRS cells (lower panels). β2M (C) and MHC class I (D) expression on bare HRS cells (cyan) and CD3+ rosetted HRS cells (dark blue) in each of the primary cHLs (1-7). Significant differences were identified using the Wilcoxon rank-sum test. (E) Dual immunohistochemical analysis (PAX5 [red] and MHC class I [brown]) of primary cHLs (cases 2, 5, and 7) and a cHL with known HRS cell expression of MHC class I (positive control [Pos.]); arrows denote HRS cell membranes.3
Using the rosetted T-cell expression of β2M and MHC class I as a frame of reference, we next compared β2M and MHC class I expression levels on bare HRS cells and HRS cell/T-cell rosettes from each primary cHL (Figure 3C-D, bare HRS cells, left and HRS cell/T-cell rosettes, right). In 5 of the 7 primary cHLs, bare HRS cells expressed significantly less β2M and MHC class I than did the rosetted T cells (Figure 3C-D). In the 3 cases with available formalin-fixed paraffin-embedded biopsy slides, we confirmed the CyTOF findings by PAX5/MHC class I dual immunohistochemistry (Figure 3E). In cases 5 and 7, HRS cells had relatively decreased cell surface MHC class I expression, whereas in case 2, HRS cells exhibited membranous MHC class I (Figure 3E).
Of interest, the 5 cHLs with relatively decreased β2M and MHC class I expression on HRS cells (Figure 3C-D, 3-7) were all EBV−, whereas the 2 cHLs with intact β2M and MHC class I expression on HRS cells (Figure 3C-D, 1 and 2) were EBV+. These findings are consistent with prior studies in which EBV+ cHLs were more likely to retain MHC class I expression compared with EBV− cHLs.13,24-26
CyTOF analyses of the inflammatory/immune cell infiltrate
Identification of discrete immune cell clusters.
After characterizing malignant HRS cells in the primary cHLs, we evaluated the inflammatory/immune cell infiltrates in these tumors. For these studies, we focused on viable singlet cells from the 7 primary cHLs and an additional 10 control RLNT samples.
From each of the malignant and control samples, 15 000 viable single cells were imported into the VorteX visualization environment and analyzed using the clustering algorithm, X-shift.21 The design of X-shift specifically allows every unique population within a complex mixture to be identified. Each cluster was labeled with a unique color based on the hex color code (supplemental Figure 2A). The identified clusters were visualized by randomly sampling a proportional number of events from each cluster and subsequently generating an FDL that included all cHL and RLNT samples (Figure 4A; supplemental Video 1).
CyTOF analyses of all viable cells. (A) FDLs generated from X-shift analysis within the VorteX visualization environment of all viable singlet cells from 7 primary cHLs and 10 RLNTs. A total of 15 000 events was collected from each sample, and the resulting 255 000 events were pooled together prior to clustering. The X-shift algorithm clusters events according to similarities in expression of CyTOF panel proteins, grouping events with shared lineage, differentiation, and polarization within the pool. Every identified unique population is labeled with a specific color based on the hex color code. (B) Major lineages delineated by expression of key markers: T cells (CD4+ and CD8+), B cells (PAX5+ and MHC class II+), NK cells (CD56+), and macrophages (PD-L1+, MHC class II+, and PAX5−). (C) Heat map of relative expression of each CyTOF panel protein marker in clusters denoted by hex color code (y-axis). Relative expression defined by a z-score. Indicated clusters contain >5% of sampled events per case. Clusters are defined by the expression of lineage and differentiation markers (left, x-axis; right, phenotype key).
CyTOF analyses of all viable cells. (A) FDLs generated from X-shift analysis within the VorteX visualization environment of all viable singlet cells from 7 primary cHLs and 10 RLNTs. A total of 15 000 events was collected from each sample, and the resulting 255 000 events were pooled together prior to clustering. The X-shift algorithm clusters events according to similarities in expression of CyTOF panel proteins, grouping events with shared lineage, differentiation, and polarization within the pool. Every identified unique population is labeled with a specific color based on the hex color code. (B) Major lineages delineated by expression of key markers: T cells (CD4+ and CD8+), B cells (PAX5+ and MHC class II+), NK cells (CD56+), and macrophages (PD-L1+, MHC class II+, and PAX5−). (C) Heat map of relative expression of each CyTOF panel protein marker in clusters denoted by hex color code (y-axis). Relative expression defined by a z-score. Indicated clusters contain >5% of sampled events per case. Clusters are defined by the expression of lineage and differentiation markers (left, x-axis; right, phenotype key).
The FDL revealed individual clusters arranged into larger groups defined by known cell subset markers: CD4, CD8, CD56, PAX5, and MHC class II (Figure 4B). The majority of viable singlet cell clusters were CD4+ or CD8+ T cells or PAX5+/MHC class II+ B cells (Figure 4A-B). Smaller clusters of CD56+ NK cells and CD3+/CD4−/CD8− cells were detected, and additional monocyte/macrophage clusters were defined by their expression of PD-L1, MHC class II, and CD68 (Figure 4; supplemental Figure 2).
To systematically assess each of the identified clusters, we generated a heat map reflecting the relative expression of each of the CyTOF panel proteins. Clusters with >5% of sampled events are shown in the heat map in Figure 4C; all clusters are included in the heat map in supplemental Figure 2B.
Analyses of T-cell differentiation.
The T-cell population was initially divided into CD4+ and CD8+ subsets and subsequently characterized as naive, central memory (CM), effector memory (EM), or terminally differentiated effector memory (TEMRA) cells using a combination of CCR7 and CD45RO (Figure 4C).27,28 Within the CD4+ population, regulatory T cells (Tregs) were classified as CD25+ and FoxP3+, and T follicular helper (TFH) cells were classified as PD-1++ (high), CXCR5+, and CCR7− (Figure 4C).29
B-cell clusters.
Several B-cell clusters were identified by the coexpression of PAX5 and MHC class II, including CD73+ memory-type cells30 and an additional Ki-67+ subset31 (Figure 4C). We also detected a small PAX5+/MHC class II+ singlet cluster that coexpressed CD30 and had lower levels of β2M and MHC class I (Figure 4A,C; Supplemental Figure 2B [ID: 8573]).
NK cells and CD3+/CD4−/CD8− T cells.
Only 1 CD56+ NK cell cluster represented >5% of sampled events (Figure 4); these cells also expressed CD161, an additional marker of NK lineage commitment (Figure 4C).32 The small subsets of CD3+/CD4−/CD8− T cells (>5% of sampled events) also expressed CD161 (supplemental Figure 2; ID: 8550 and 8520). Such CD161+/CD3+/CD4−/CD8− cells may represent γ/δ or mucosal-associated invariant T cells, which recognize non–MHC class I–presented antigens.33,34
Macrophages.
Myeloid cells and macrophages were underrepresented in the analysis, potentially due to the fragility and adhesiveness of these cells. Among the macrophage populations, we detected a small MHC class II+/PD-L1+/CD68+/CD163−/IRF4+ cluster (ID: 8514) and an additional MHC class II+/PD-L1+/CD68+/CD163+/IRF4+ cluster (ID: 8543) (supplemental Figure 2B-C). These 2 distinct clusters, which differ in their CD163 expression, may reflect M1 and M2 polarization.35,36
CD3+ T cells.
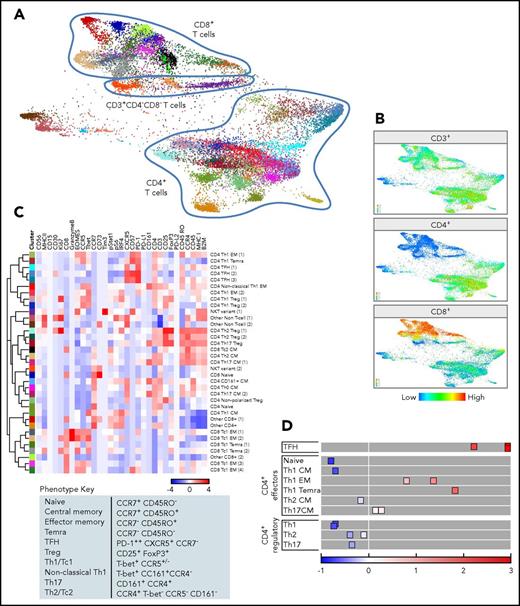
To further define the distinct T-cell subsets, we performed an additional X-shift analysis that was restricted to the viable CD3+ singlet population (Figure 5A). The FDL visualizes individual clusters arranged into larger groups of CD8+ or CD4+ cells and a smaller group of CD3+/CD4−/CD8− cells (Figure 5A-B; supplemental Figure 3A).
CyTOF analyses of CD3+cells. (A) FDLs generated from X-shift within VorteX visualization environment of CD3+ cells from 7 primary cHLs and 10 RLNTs. A total of 7350 events was collected from each sample, and the resulting 124 950 events were pooled together prior to clustering. The X-shift algorithm clusters events according to similarities in expression of CyTOF panel proteins, grouping events with shared lineage, differentiation, and polarization within the pool. Every identified unique population is labeled with a specific color based on the hex color code. (B) CD4+, CD8+, and CD3+/CD4−/CD8− T-cell clusters defined by the relative expression of CD4+, CD8+, or neither marker. (C) Heat map of relative expression of the indicated CyTOF panel proteins in clusters denoted by hex color code (y-axis). Relative expression defined by a z-score. Indicated clusters contain >5% of sampled events per case. Clusters are defined by the expression of lineage, differentiation, polarization, and function markers (top, x-axis; bottom, phenotype key). (D) Graphical representation of relative PD-1 levels in CD4+ cell subsets. The 3 TFH subsets express the highest levels of PD-1, and naive cells do not express PD-1. By comparison, Tregs are negative for PD-1, and Th1-polarized effector cells (EM cells and TEMRA cells) have increased PD-1 expression.
CyTOF analyses of CD3+cells. (A) FDLs generated from X-shift within VorteX visualization environment of CD3+ cells from 7 primary cHLs and 10 RLNTs. A total of 7350 events was collected from each sample, and the resulting 124 950 events were pooled together prior to clustering. The X-shift algorithm clusters events according to similarities in expression of CyTOF panel proteins, grouping events with shared lineage, differentiation, and polarization within the pool. Every identified unique population is labeled with a specific color based on the hex color code. (B) CD4+, CD8+, and CD3+/CD4−/CD8− T-cell clusters defined by the relative expression of CD4+, CD8+, or neither marker. (C) Heat map of relative expression of the indicated CyTOF panel proteins in clusters denoted by hex color code (y-axis). Relative expression defined by a z-score. Indicated clusters contain >5% of sampled events per case. Clusters are defined by the expression of lineage, differentiation, polarization, and function markers (top, x-axis; bottom, phenotype key). (D) Graphical representation of relative PD-1 levels in CD4+ cell subsets. The 3 TFH subsets express the highest levels of PD-1, and naive cells do not express PD-1. By comparison, Tregs are negative for PD-1, and Th1-polarized effector cells (EM cells and TEMRA cells) have increased PD-1 expression.
Identification of polarized Teffs and Tregs.
We next generated heat maps for CD3+ clusters (>5% of sampled events, Figure 5C; all CD3+ clusters, supplemental Figure 3B) and assigned an initial lineage and differentiation phenotype to each cluster using the above-mentioned criteria (Figure 4C). In addition, we used a combination of T-bet, CCR5, CCR4, and CD161 expression to define nonpolarized and polarized effector subsets: CD8+ Tc1 cells, Tc2 cells, and CD4+ T helper 1 (Th1), nonclassical Th1, Th17, and Th2 cells (Figure 5C). With these markers, we also identified polarized Treg subsets (Th1, Th2, and Th17 Tregs), in addition to their T effector (Teff) counterparts.37,38
PD-1 expression on polarized Teffs and Tregs.
After characterizing lineage, differentiation, and polarization, we assessed the functional status of specific T-cell subsets using markers of activation and exhaustion, including PD-1 (Figure 5C-D; supplemental Figure 3B). As expected, CD4+ TFH cells expressed the highest levels of PD-1 (Figure 5C-D).29 The more differentiated and polarized CD4+ Th1 EM and TEMRA cells also expressed PD-1 (Figure 5C-D). In contrast, the less differentiated and polarized CD4+ Th1, Th2, and Th17 CM cells had lower levels of PD-1 expression, and CD4+ naive cells were PD-1− (Figure 5C-D). Of interest, polarized CD4+ Th1, Th2, and Th17 Tregs had low or no PD-1 expression (Figure 5C-D).
NK T cells.
In the CD3+ FDLs (Figure 5A; supplemental Figure 3A), we also identified 3 small CD4−/CD8− clusters of likely NK T cells, including CD56dim/CD57+/CD161− cells (ID: 18847), CD56high/CD57−/CD161+ cells (ID: 18848, <5% of sampled events), and CD56dim/CD57−/CD161− cells (ID: 18853) (supplemental Figure 3B). In such cells, the combination of low-level CD56 expression and CD57 positivity likely defines an activated population (ID: 18847).39,40
Comparative analyses of the inflammatory/immune cell infiltrate in cHLs and RLNTs
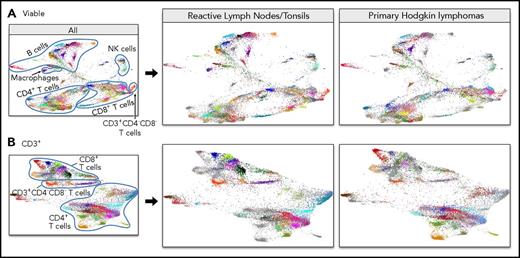
The initial FDLs (Figures 4A, 5A) included events from primary cHLs and RLNTs. To distinguish cHL-associated immune changes from normal secondary lymphoid organ infiltrates, we separated the FDL of cHL samples from that of RLNTs (Figure 6). In these separated FDLs (all viable cells, Figure 6A; CD3+ cells, Figure 6B), events pertaining to the group of interest retain their hex color code (supplemental Figures 2A, 3A), whereas events belonging to the other group are represented in gray.
Separate FDLs of RLNTs and primary cHLs. All viable cell clusters (A) and CD3+ T-cell clusters (B). In each FDL, the events pertaining to the group of interest retain their hex color code. Events belonging to the other group are represented in gray.
Separate FDLs of RLNTs and primary cHLs. All viable cell clusters (A) and CD3+ T-cell clusters (B). In each FDL, the events pertaining to the group of interest retain their hex color code. Events belonging to the other group are represented in gray.
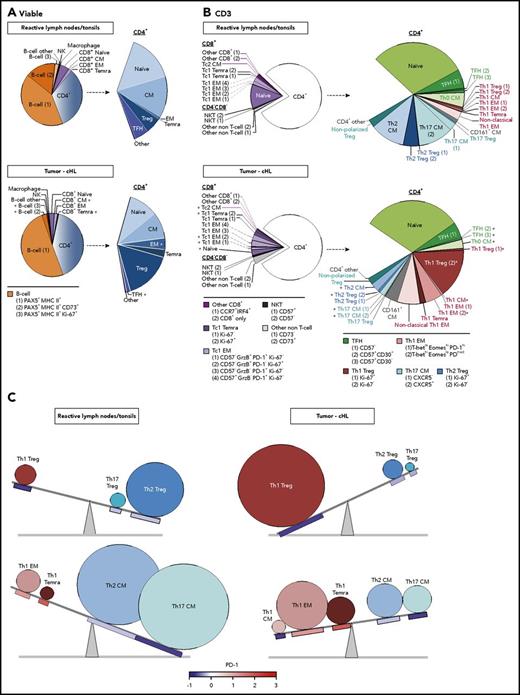
There were notable differences in the representation and abundance of specific singlet clusters in the RLNT and primary cHL cell suspensions (Figure 6). To quantify these differences, we determined the number of cells that each RLNT sample and cHL specimen contributed to a given cluster and to shared categories with a common lineage, differentiation, and polarization status (as in Figure 5C). For each cluster and shared category, the median RLNT and cHL cell counts were displayed as pie charts (Figure 7A, all viable cells; Figure 7B, CD3+ cells). Additional distinguishing features were added to further characterize the identified B-cell and CD3+ clusters (Figure 7A-B, bottom). A minority of clusters did not have a readily identifiable phenotype and were classified as “other” (Figure 7A-B). For the individual clusters and the clusters with shared lineage, differentiation, and polarization status, we performed a Wilcoxon rank-sum test to identify significant differences in abundance in cHLs vs RLNTs (Figure 7; supplemental Figure 4).
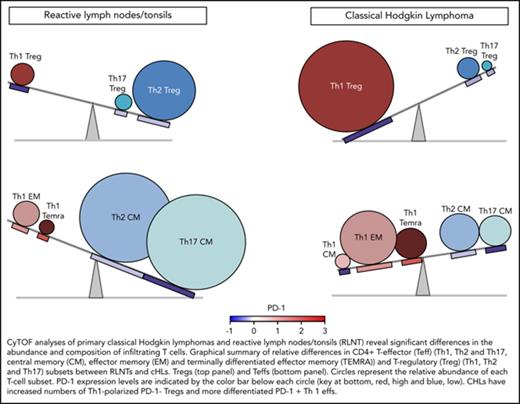
Comparative analyses of the immune cell infiltrates in RLNTs and primary cHLs. To quantify differences between RLNTs and cHL specimens, the number of cells that each contributed to a given cluster was determined. Clusters were defined by lineage, differentiation, and polarization status. For each cluster and shared category, the RLNT and cHL median cell count was calculated and presented as comparison pie charts. (A) All viable cell clusters, >5% of sampled events per cases. (B) CD3+ T cell clusters, >5% of sampled events per case. Clusters that are statistically significantly different in cHLs and RLNTs are marked with an asterisk (*; see supplemental Figure 4). (C) Relative differences in CD4+ Teff (lower panels) and Treg (upper panels) subsets between RLNTs and cHLs. The relative proportions of each subset are represented by the sizes of the circles, and the color scheme is the same as in (B). PD-1 expression levels for each CD4+ T-cell subset are indicated below the respective circle and are based on the color bar shown at the bottom of the panel. cHLs have increased numbers of Th1-polarized PD-1− Tregs and more differentiated PD-1+ Th1 effector cells.
Comparative analyses of the immune cell infiltrates in RLNTs and primary cHLs. To quantify differences between RLNTs and cHL specimens, the number of cells that each contributed to a given cluster was determined. Clusters were defined by lineage, differentiation, and polarization status. For each cluster and shared category, the RLNT and cHL median cell count was calculated and presented as comparison pie charts. (A) All viable cell clusters, >5% of sampled events per cases. (B) CD3+ T cell clusters, >5% of sampled events per case. Clusters that are statistically significantly different in cHLs and RLNTs are marked with an asterisk (*; see supplemental Figure 4). (C) Relative differences in CD4+ Teff (lower panels) and Treg (upper panels) subsets between RLNTs and cHLs. The relative proportions of each subset are represented by the sizes of the circles, and the color scheme is the same as in (B). PD-1 expression levels for each CD4+ T-cell subset are indicated below the respective circle and are based on the color bar shown at the bottom of the panel. cHLs have increased numbers of Th1-polarized PD-1− Tregs and more differentiated PD-1+ Th1 effector cells.
Comparative analyses of all viable cell groups in cHLs and RLNTs
The median numbers of total CD8+ and CD4+ T cells in the RLNT and cHL samples were not significantly different (Figure 7A; supplemental Figure 4A). In both types of samples, CD4+ cells were more than fivefold more abundant than CD8+ cells (Figure 7A; supplemental Figure 4A). Although total B-cell numbers were comparable in cHLs and controls, more specifically defined B-cell clusters, such as CD73+ memory B cells, were less abundant in cHLs (P = .002) (Figure 7A; supplemental Figure 4A).
Skewed T-cell differentiation in cHLs
Although the percentages of total CD8+ and CD4+ T cells were comparable in cHLs and RLNTs, the cHL CD8+ and CD4+ T-cell infiltrates were significantly more differentiated. Primary cHLs included relatively fewer CD8+ CM cells (P = .005) and more terminally differentiated CD8+ TEMRA cells (P = .004) (Figure 7A; supplemental Figure 4A). These tumors also had more abundant CD4+ EM cells (P = .014) and CD4+ TEMRA cells (P = .07) (Figure 7A; supplemental Figure 4A). In contrast, CD4+ TFH cells were significantly decreased in cHLs (P = .007) (Figure 7A; supplemental Figure 4A). As CD4+ TFH cells support the growth and differentiation of memory B cells,30 this B-cell subset may be less abundant in primary cHLs because of their relative paucity of TFH cells (Figure 7A; supplemental Figure 4A).
Increased terminal differentiation and Tc1 polarization in cHL
We next assessed potential differences in polarized T-cell subsets in cHL and RLNT samples, using CD3+ FDLs (Figure 6B), associated pie charts (Figure 7B), and statistical comparisons (supplemental Figure 4B). In cHLs, the more terminally differentiated CD8+ cells were also more Tc1 polarized (Figure 7B; supplemental Figure 4B). Specifically, CD8+ naive cells were less abundant (P = .014) and granzyme B+ Tc1 EM cells and Ki-67+ Tc1 TEMRA cells were more abundant in cHLs [Tc1 EM (1), P = .017; Tc1 EM (2), P = .012; Tc1 TEMRA cells (2), P = .019] (Figure 7B; supplemental Figure 4B).
Although EBV+ and EBV− cHLs had increased numbers of granzyme B+ Tc1 EM cells, only EBV+ cHLs had more abundant Tc1 TEMRA cells (supplemental Figures 5 and 6). The increased terminal differentiation of Tc1 cells in EBV+ cHLs may reflect the intact MHC class I–mediated antigen presentation in these tumors (Figure 3). In EBV+ and EBV− cHLs, CD8+ Tc2 cells were comparatively less differentiated (ie, only Tc2 CM cells with no EM cells or TEMRA cells) and less frequent (P = .005) (Figure 7B; supplemental Figures 5 and 6).
Increased terminal differentiation and Th1 polarization in cHL
In EBV+ and EBV− cHLs, the CD4+ T-cell infiltrate was also more terminally differentiated and Th1 polarized than that in RLNTs (supplemental Figures 5 and 6). In all cHLs, CD4+ Th0 CM cells were less abundant (P = .001), whereas CD4+ Th1 CM, EM (2), and TEMRA cells were relatively more frequent (P ≤ .001, P = .015, and P = .097, respectively) (Figure 7B; supplemental Figure 4B).
Increased active Th1 Tregs and exhausted terminally differentiated Th1 Teffs in cHL
In addition to having expanded numbers of differentiated CD4+ Th1-polarized Teffs, the primary cHLs contained significantly more abundant Th1-polarized Tregs (Ki-67−, P = .007; Ki-67+, P = .002) (Figure 7B; supplemental Figure 4B). Of note, the expanded CD4+ Th1 Treg population was PD-1−, whereas the CD4+ Th1 EM cells and TEMRA cells were PD-1+ (Figure 5D). These findings highlight 2 complementary bases of CD4+ Th1-dependent immune evasion in cHLs: likely active Th1 Tregs and exhausted Th1 EM cells (Figure 5D). In addition, the expanded CD4+ Th1 EM (2) cells in cHL have the phenotype T-bethigh EOMESlow PD-1medium, which is associated with enhanced susceptibility to PD-1 blockade (Figure 7B).41
In contrast to the more abundant CD4+ Th1 Teffs and Tregs, CD4+ Th2 CM cells and Tregs were less abundant in these primary cHLs (Th2 CM, P = .003; Th2 Tregs [all], P = .01) (Figure 7B; supplemental Figure 4B). Furthermore, the major CXCR5+ and minor CXCR5− Th17 CM cell subsets were less abundant in the primary cHLs than in RLNTs (P = .019 and P = .01, respectively) (Figure 7B; supplemental Figure 4B).
Taken together, these data define a more terminally differentiated CD4+ T-cell predominant and Th1-polarized immunosuppressive microenvironment in cHL (Figure 7C).
Discussion
In this study, a comprehensive CyTOF analysis of the cHL TME revealed decreased β2M and MHC class I expression on individual HRS cells in the majority of cHLs (Figure 3), a concomitant increase in CD4+ Th1-polarized Tregs and differentiated Teffs in primary cHL suspensions (Figure 7C), and complementary bases of immunosuppression in primary cHLs: exhausted PD-1+ Th1 Teffs and likely active PD-1− Th1 Tregs (Figure 7C).
The observed loss of β2M and MHC class I expression on HRS cells in the majority of cases is consistent with previous reports.3,12,13 In the current small series, cHLs with intact MHC class I–mediated antigen presentation had increased numbers of Tc1-polarized terminally differentiated cytotoxic CD8+ T cells. However, all of the primary cHLs had expanded numbers of CD4+ Th1-polarized Tregs and Teffs, irrespective of HRS cell MHC class I status.
Our detailed analyses build upon prior descriptions of a CD4+ T-cell predominant Treg-rich TME in cHL.19,42-45 With the additional markers that capture polarization and functional status and delineate effector and regulatory subsets, we identified selective expansion of more differentiated CD4+ Th1-polarized Teffs (EM and TEMRA cells) and associated Th1-polarized Tregs in cHL cell suspensions (Figure 7C). These findings likely reflect the known proinflammatory TME in cHL and the polarization of Tc1/Th1 Teffs and Th1 Tregs by the same predominant Th1 transcription factors.38,46,47 Of interest, recent studies indicate that intratumoral Tregs are more likely to express recurrent αβ T-cell receptors, suggestive of specific antigen exposure.48,49 Consistent with these observations, the expanded Th1 Treg population in the cHLs has features (CCR7low Ki-67low) associated with a memory phenotype50 (Figures 5C and 7B).
By simultaneously assessing PD-1 levels on polarized CD4 Teffs and Tregs in cHL, we found that the expanded differentiated Th1 EM cells and TEMRA cells expressed intermediate and high levels of PD-1, whereas the Th1 Tregs were PD-1low/negative (Figure 5D). In our cHL tumor cell suspensions, the most significantly expanded CD4+ Th1 EM population was T-bethigh EOMESlow PD-1medium (Figure 7B). Although the comprehensive signatures of exhausted CD4+ subsets are less well characterized than those of CD8+ effector cells, these cells share major core transcriptional modules.51 T-bethigh EOMESlow PD-1medium effector cells are reported to be most amenable to PD-1 blockade,41,52,53 of note given the efficacy of this approach in cHL.
The relative levels of PD-1 expression and consequences of PD-1 signaling in Tregs are less well defined. However, in other tumors, the relative absence of PD-1 has been associated with functionally active Tregs.54,55 As a consequence, our CyTOF analyses reveal potentially complementary and targetable mechanisms of CD4 T-cell–dependent immunosuppression in cHL: Th1-polarized functionally active PD-1− Tregs and differentiated and likely exhausted PD-1+ effector cells (Figure 7C).
Our detailed characterization of the TME of primary cHL cell suspensions sets the stage for subsequent comparative analyses of newly diagnosed and relapsed cHLs and detailed characterization of the immune response to PD-1 blockade, including the potential role of additional non–MHC class I–restricted effector cells.56,57 It will also be possible to build upon the current approach, using an analogous CyTOF panel and multiplexed ion beam imaging,58 to evaluate the intact cHL TME in further detail.
In conclusion, the current analysis provides new insights into the complex cHL TME in which CD4+ Th1-polarized Tregs and Teffs with different levels of PD-1 expression promote immune evasion.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ana Lako and Sarah Abdelrahman for technical assistance.
This work was supported in part by Bloodwise 14042 (F.Z.C.) and by National Institutes of Health, National Cancer Institute grant R01 CA161026 (M.A.S.), the International Immune Oncology Network of Bristol-Myers Squibb (M.A.S. and S.J.R.), the Miller Family Fund (M.A.S.), and National Institutes of Health, National Cancer Institute grant U24CA224316 (X.S.L. and X.H.). F.Z.C. was the recipient of a Helen Gurley Brown Fellowship.
Authorship
Contribution: F.Z.C. designed and performed research, analyzed data, and wrote the manuscript; R.C.J.S., X.H., X.S.L., R.R., and D.N. analyzed data; K.W., S.J.R., D.W., and J.R.F. performed research and analyzed data; B.C., J.O., and P.A. provided critical feedback and helped to shape the research and analysis; N.P. acquired CyTOF data; E.G. and M.L. performed experiments; and M.A.S. designed and supervised the research and data analyses and wrote the manuscript. All authors discussed the results and contributed to the final version of the manuscript.
Conflict-of-interest disclosure: M.A.S. has received research funding from BMS and served on advisory boards for BMS and Merck. P.A. consults for BMS, Merck, Pfizer, and Infinity and receives institutional research funding from BMS, Merck, Affimed, Sequenta, Roche, Tensha, Otsuk, and Sigma-Tau. The remaining authors declare no competing financial interests.
Correspondence: Margaret A. Shipp, Dana-Farber Cancer Institute, Harvard Medical School, 450 Brookline Ave, Mayer 513, Boston, MA 02215; e-mail: margaret_shipp@dfci.harvard.edu.
REFERENCES
Author notes
X.H., K.W., and R.R. contributed equally to this work.



![Figure 3. Analysis of β2M and MHC class I expression on HRS cells by CyTOF. (A) Rosetted HRS cells separated from bare HRS cells by sequential gates. Events from the nonsinglet population are split according to CD3+ expression. Events within the CD3− gate are further gated to identify the CD4−/CD8− population. The CD3−/CD4−/CD8− subset was positively gated to select cells that coexpressed CD30 and CD15, representing bare HRS cells. Within the CD3+ population, CD15+/CD30+ coexpression identifies rosetted HRS cells. (B) CD3+ expression on rosetted HRS cells (upper panels) and bare HRS cells (lower panels). β2M (C) and MHC class I (D) expression on bare HRS cells (cyan) and CD3+ rosetted HRS cells (dark blue) in each of the primary cHLs (1-7). Significant differences were identified using the Wilcoxon rank-sum test. (E) Dual immunohistochemical analysis (PAX5 [red] and MHC class I [brown]) of primary cHLs (cases 2, 5, and 7) and a cHL with known HRS cell expression of MHC class I (positive control [Pos.]); arrows denote HRS cell membranes.3](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/8/10.1182_blood-2018-04-843714/4/m_blood843714f3.jpeg?Expires=1764993987&Signature=rdN3wShgJKGBx5nJh4T8zLBgotzGzRubNfG71XpPFQa7R3YvSri98bUqfWS2axtNKs1cvtUII9PmmDdGiE8X1u~5bwzhtjF33CNpV20v6kbwPemljuFdZa4YO-GaVYrWNGsuZxTNm4xeaoXP6-PuckiBLd86BrRYKX0ETBhHKM95N8m6-KjAAx1FluZYW9RVrCyP2v8NlnFqOKiquyHBc8AO-q53g-mxskNg7k2MYn3lHE4ioTKSAx~OpOb-2Gp34bknuZqE5UNX6auzuCr0Id0xas~YjMrZoIG3fhANx~Vg1y9hF87exL1VWIcaBv9Q7Wr5ZjYf79k942nDrbFBWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal