Key Points
First quantitative analysis of dynamic platelet lipidome modulation reveals key lipids altered in platelet activation.
Lipidomics in a knockout approach unravel SMPD1 as a powerful modulator of platelet lipidome and activation via regulation of SPC.
Abstract
Platelet integrity and function critically depend on lipid composition. However, the lipid inventory in platelets was hitherto not quantified. Here, we examined the lipidome of murine platelets using lipid-category tailored protocols on a quantitative lipidomics platform. We could show that the platelet lipidome comprises almost 400 lipid species and covers a concentration range of 7 orders of magnitude. A systematic comparison of the lipidomics network in resting and activated murine platelets, validated in human platelets, revealed that <20% of the platelet lipidome is changed upon activation, involving mainly lipids containing arachidonic acid. Sphingomyelin phosphodiesterase-1 (Smpd1) deficiency resulted in a very specific modulation of the platelet lipidome with an order of magnitude upregulation of lysosphingomyelin (SPC), and subsequent modification of platelet activation and thrombus formation. In conclusion, this first comprehensive quantitative lipidomic analysis of platelets sheds light on novel mechanisms important for platelet function, and has therefore the potential to open novel diagnostic and therapeutic opportunities.
Introduction
Platelet adhesion and activation are essential for primary hemostasis and critical for the development of acute thrombotic occlusion, the major pathophysiological mechanism underlying myocardial infarction or ischemic stroke.1 Platelets are activated by several prothrombotic mediators, including subendothelial collagen and thrombin.2 After the stimulation of protease-activated receptors (PARs) or glycoprotein VI (GPVI), platelets become activated, secrete their granules following fusion with the plasma membrane (PM), and undergo cell-membrane scrambling.3 The latter leads to phosphatidylserine (PS) exposure at the outer leaflet of the PM, with entry into a procoagulant platelet state.4,5 For degranulation and membrane scrambling, rearrangement of membrane lipid composition and fluidity is essential. Degranulation and membrane scrambling in platelets and other cells involve sphingomyelin (SM) phosphodiesterase-1 (Smpd1).6,7 Smpd1 breaks down the sphingolipid (SL) SM, which is a significant part of the platelet PM.8 Ceramides (Cers) thus produced from Cer-rich platforms modulating granule fusion and fission by affecting composition and curvature of the PM. SM hydrolysis is critical for platelet membrane properties because platelets lack de novo synthesis of Cers.9 Excessive Cer formation contributes to a wide variety of disorders, including atherosclerosis,10,11 cerebral ischemia,12 and cardiovascular diseases,13 which may all result from dysregulated platelet function.
Platelets are major targets for the treatment of thrombo-occlusive disorders. However, key challenges remain because many patients suffering from acute arterial thrombotic events do not respond well to existing clinical treatments and/or suffer from severe side effects. Lipids are essential for platelet integrity and play a fundamental role for platelet lifespan and senescence, as well as for platelet shape change and aggregation.14-18 Various lipids have been described in resting and activated platelets.19,20 In the first global study, 5000 putative lipid species were detected indicating the high complexity of the platelet lipidome.19 The study further shed light on the functional significance of the lipidome such as regulation of mitochondrial bioenergetics by PLA2.19 Nevertheless, no comprehensive study sequencing and describing the platelet lipidome from a quantitative perspective is available. Knowledge of concentrations is essential for comparing lipids between species or cross species, and to fully understand the mechanisms underlying initiation and propagation of platelet activation. Quantification of membrane and signaling lipids is clearly required for correct modeling of membrane fluidity, stiffness, or signaling.21
For a full structural analysis of the lipids, we developed the first comprehensive quantitative lipidomics analysis tailored for murine platelets by providing lipid concentrations of 384 lipid species, spanning 28 lipid classes, monitored in resting and activated platelets. Key findings were confirmed in human platelets. The method was further validated by the analysis of a rare lysosomal lipid storage disease caused by a point mutation of the SMPD1 (acidic sphingomyelinase) gene-on-gene locus 11p15.1-15.4.22 The mutation causes a reduced expression of the active enzyme.23 We analyzed the impact of SMPD1 on the platelet lipidome in resting and activated platelets isolated from mice with a genetic knockout of Smpd1 (Smpd1−/−) and wild-type littermates (Smpd1+/+).
Methods
Chemicals
A detailed description of the chemicals used is provided in supplemental Experimental procedures, available on the Blood Web site.
Platelet isolation and stimulation
Platelets were obtained from C57BL/6 wild-type mice (Smpd1+/+) and littermates with a genetic knockout of acid sphingomyelinase (Smpd1−/−) or isolated from human volunteers, freshly isolated and stimulated as described in detail in the supplemental Experimental procedures.
Lipid extraction
Lipid extraction (except for lipid mediators [LMs]) was carried out according to Matyash et al.24 The dried extract was resuspended for further mass spectrometer (MS) analysis. To extract LMs, acetic acid was additional added. In total, 36 internal standards were used for quantification. A detailed description of lipid extraction and standards is reported in the supplemental Experimental procedures.
Lipid analysis
Shotgun analysis was performed using a NanoMate coupled to a Q-ExactivePlus. All spectra were analyzed by LipidXplorer. Targeted analyses of SLs and LMs were performed using liquid chromatography (LC) coupled to QTRAP6500. Data analysis including peak integration and chromatogram quality control was carried out using Skyline. Separation, MS acquisition, and data analysis parameters are detailed in the supplemental Experimental procedures.
Visualization and network analysis
A violin plot was selected to show the distribution of the abundances per class. Network analysis was performed using Cytoscape to show correlations and anticorrelations with a Pearson correlation coefficient value above 0.85 (supplemental Experimental procedures).
Analysis of platelet activation and in vitro thrombus formation
Lysosphingomyelin (SPC) was dissolved and stored according to the manufacturer’s guidelines. Freshly isolated platelets or whole blood were preincubated for 2 minute25 at room temperature with the indicated concentrations of SPC. Platelet secretion (α and dense granules), aggregation and in vitro thrombus formation were analyzed using luminescence aggregometry, fluorescence-activated cell sorter analysis, and collagen-coated flow chamber as described previously26 and detailed in supplemental Experimental procedures.
Results
Comprehensive quantitative lipidomics analysis
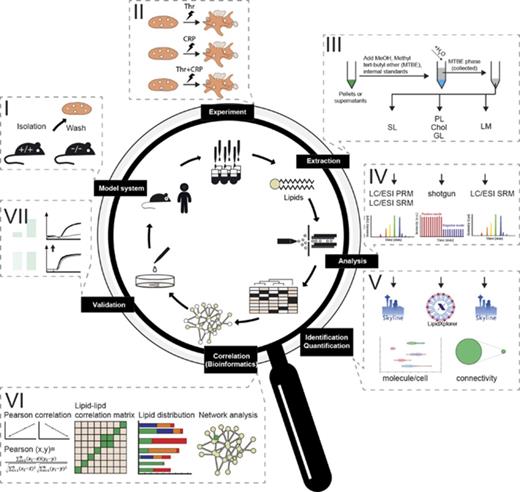
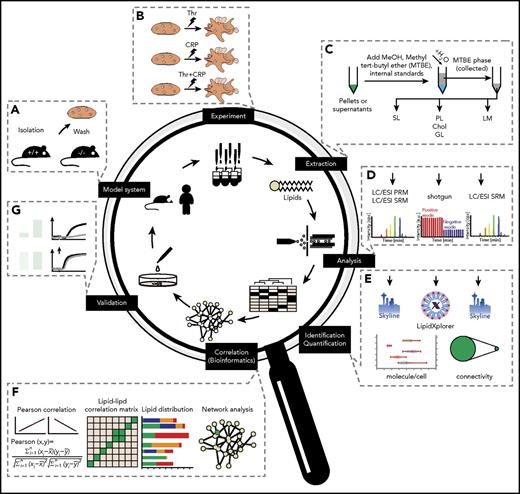
For global quantitative platelet-lipidomics assay allowing simultaneous detection of low-abundance lipid species (eicosanoids) along with major membrane components (cholesterol) from a single mouse species, we established a workflow (Figure 1) integrating top-down shotgun and targeted lipidomics.27,28 Platelets were washed, stimulated, perturbed, and extracted with a lipid-category tailored extraction procedure. For high accuracy and absolute quantification, the analysis used internal lipid standards at species or lipid-class level.27,29 After structural analysis, identification, and quantification, the absolute amounts obtained were subjected to Pearson correlation, followed by cosine similarity clustering taking positive and negative correlation into account, and were finally used for network analysis. Identified changes were validated in humans and major findings were investigated by ex vivo experiments; the predicted effects on platelet activation were confirmed.
Comprehensive lipidomic analysis for the absolute quantification of the platelet lipidome. (A) Platelet isolation and purification from mice and human models. (B) Ex vivo stimulation using different stimuli. (C) Lipid extraction and internal standard addition applying to lipid-category tailored analysis. (D) Lipid analysis by shotgun or targeted lipidomic analysis. (E) Data analysis and structural characterization by LipidXplorer or Skyline, followed by absolute quantification of all detected lipid species. (F) Establishment of a correlation and anticorrelation lipid-lipid matrix to form a lipid network. Elucidation of knockout (KO) models and perturbation-specific differences identified from absolute quantification of lipids. (G) Validation by additional ex vivo experiments, including functional assays.
Comprehensive lipidomic analysis for the absolute quantification of the platelet lipidome. (A) Platelet isolation and purification from mice and human models. (B) Ex vivo stimulation using different stimuli. (C) Lipid extraction and internal standard addition applying to lipid-category tailored analysis. (D) Lipid analysis by shotgun or targeted lipidomic analysis. (E) Data analysis and structural characterization by LipidXplorer or Skyline, followed by absolute quantification of all detected lipid species. (F) Establishment of a correlation and anticorrelation lipid-lipid matrix to form a lipid network. Elucidation of knockout (KO) models and perturbation-specific differences identified from absolute quantification of lipids. (G) Validation by additional ex vivo experiments, including functional assays.
Lipidome of resting platelets
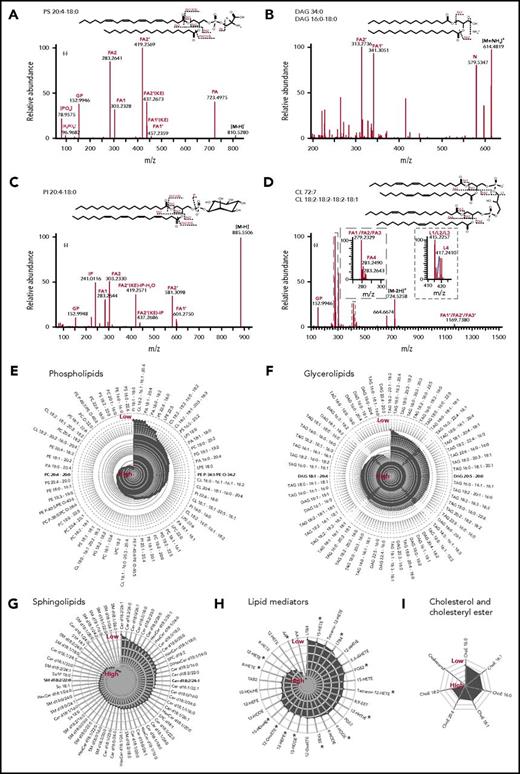
For the high-confidence identification of lipids, all lipid molecules were structurally characterized by tandem MS analysis (MS/MS).30-32 Glycerolipids (GLs) and glycerophospholipids (PLs) were analyzed by direct infusion using high-resolution MS (HR-MS). Cholesteryl ester (ChoE), cholesterol, and low-abundance lipid species, such as LMs and SLs, were analyzed by reversed-phase LC coupled to a triple quadrupole MS or HR-MS. Thus, PLs and GLs were identified by accurate mass and at least 3 specific fragment ions (Figure 2A-D). SLs were identified by exact mass and the coeluting fragmentation pattern (supplemental Figure 1). All LMs were identified by coelution with synthetic standards or isotope-coded standards and their representative fragment patterns (supplemental Figure 2). The procedure allowed structural characterization of lipids at the fatty acid (FA) species level with high confidence, with definition of the number of double bonds and carbon atoms for each FA chain (Figure 2A-D). This information is of critical importance because major remodeling and replacement of FAs occurs during fundamental cell-fate decision processes such as apoptosis33 and senescence.34 The lipid FA composition contributes to chemicophysical properties of the membrane, including lateral diffusion and stiffness. The present analysis structurally characterized and identified 384 lipid species (Figure 2E-I) originating from the main lipid categories PL (167), GL (120), SL (60), LM (30), and Chol (7), thereby covering 28 different lipid classes (supplemental Table 1).
Structural elucidation at the FA level of lipids in the main platelet lipid categories. (A-D) Molecular structural analysis of lipids by shotgun lipidomics at the precursor and fragment level. The different panels showing different MS2 spectra that are used for structural assessment by customized mfql search files, using the fully automated search engine LipidXplorer. Potential fragmentation mechanisms are shown above each panel. Annotated fragments are denoted in the chemical structure of individual species. (E-I) Radar plots displaying the relative intensity of lipid species in resting platelets. The lipid species are presented according to their specific categories, including phospholipids (PL), GL, SL, FA, and derivatives (LM), and cholesterol and derivatives (Chol). PLs and GLs were analyzed by shotgun lipidomics and Chol, SL, and LM were analyzed by targeted lipidomics. All species were analyzed at least in 4 different independent biological experiments. DAG, diacylglycerol; TAG, triacylglycerol.
Structural elucidation at the FA level of lipids in the main platelet lipid categories. (A-D) Molecular structural analysis of lipids by shotgun lipidomics at the precursor and fragment level. The different panels showing different MS2 spectra that are used for structural assessment by customized mfql search files, using the fully automated search engine LipidXplorer. Potential fragmentation mechanisms are shown above each panel. Annotated fragments are denoted in the chemical structure of individual species. (E-I) Radar plots displaying the relative intensity of lipid species in resting platelets. The lipid species are presented according to their specific categories, including phospholipids (PL), GL, SL, FA, and derivatives (LM), and cholesterol and derivatives (Chol). PLs and GLs were analyzed by shotgun lipidomics and Chol, SL, and LM were analyzed by targeted lipidomics. All species were analyzed at least in 4 different independent biological experiments. DAG, diacylglycerol; TAG, triacylglycerol.
Quantitative investigation of the platelet lipidome
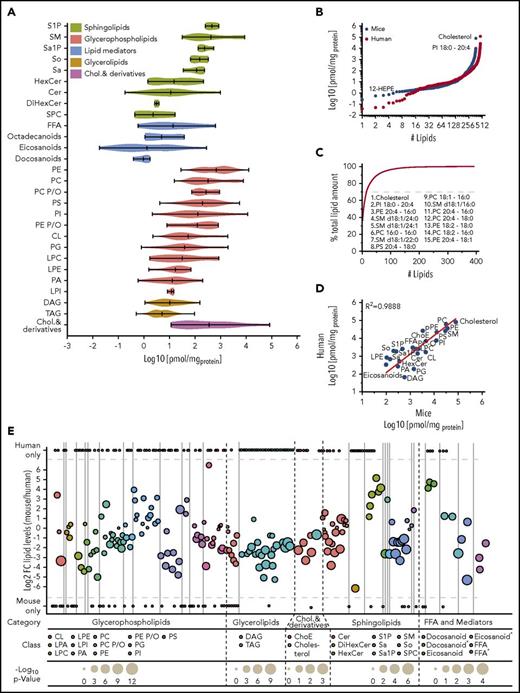
Lipids were quantified based on internal standards, a fixed number of platelets, yielding the corresponding concentrations and numbers of molecules per platelet (Figure 3A). Abundant lipids were analyzed with shotgun lipidomics yielding limit of detection/limit of quantification (LOD/LOQ) between 10 and 100/65 and 1000 fmol of main glycerol and PL classes. Low abundant lipids were detected with LC/MS yielding LODs/LOQs between 0.06 and 0.3/0.2 and 1.0 fmol for main SL classes.27,35 By aggregating the quantitative results, a dynamic range of 7 orders of magnitude (Figure 3A-B) was achieved by our approach for murine and human platelets, demonstrating that the platelet lipidome has a dynamic range similar to the proteome range in complex cells.36 Such a wide dynamic range has not been reported for any lipidome so far. In the mouse study, low-abundance lipids such as 12-HEPE (18.4 pmol/mg protein) were detected along with high-abundance species such as cholesterol (79 658 pmol/mg protein) and phosphatidylinositol (PI) 18:0-20:4 (9972 pmol/mg protein) (Figure 3B). Most of the lipid classes span a dynamic range of over 2 orders of magnitude (eg, phosphatidylcholine [PC], phosphatidylethanolamine [PE], PS, phosphatidylglycerol [PG], SM, and cardiolipin [CL]), whereas some reach over 4 orders of magnitude (eg, Cer and free fatty acyl [FFA]), or are limited to a very narrow range, such as plasmanyl/plasmalogen-PC (PC P/O) (Figure 3A). This distribution most likely reflects how often lipids are implemented in different membrane compartments or processes. Notably, only 15 lipids account for up to 70% of the entire platelet lipid mass (Figure 3C) and are therefore essential building blocks for the membrane integrity of the resting murine and human platelets. The most abundant lipid classes detected within resting platelets were cholesterol, PE, PC, SM, PS, and PI. Interestingly, even if it is known that arachidonic acid (AA) plays a central role in platelets,37,38 compared with erythrocytes in all PL classes (supplemental Figure 3), high amounts of incorporated, covalently bound AA were observed, reflecting the presence of potential precursors for mediator production upon platelet stimulation.
Unexpected dynamic range revealed through the quantitative assessment of the platelet lipidome. (A) Violin plot displaying the dynamic range of different lipid classes in a resting platelet. Each column assembles all of the quantified lipid species for 1 class. (B) Dynamic range of identified and quantified lipids species, covering 7 orders of magnitude in both human and mouse. (C) Fifteen lipids contribute 70% of the absolute lipid membrane mass of a resting platelet. (D) Correlation of presented mouse lipidome and human lipidome based on literature (supplemental Table 2) with R2 = 0.9888. (E) Quantitative comparison of the mouse and human lipidome in resting platelets at the lipid species level with a color-coding at lipid class level. All data are combined from at least 3 independent experiments, and mean values are shown.
Unexpected dynamic range revealed through the quantitative assessment of the platelet lipidome. (A) Violin plot displaying the dynamic range of different lipid classes in a resting platelet. Each column assembles all of the quantified lipid species for 1 class. (B) Dynamic range of identified and quantified lipids species, covering 7 orders of magnitude in both human and mouse. (C) Fifteen lipids contribute 70% of the absolute lipid membrane mass of a resting platelet. (D) Correlation of presented mouse lipidome and human lipidome based on literature (supplemental Table 2) with R2 = 0.9888. (E) Quantitative comparison of the mouse and human lipidome in resting platelets at the lipid species level with a color-coding at lipid class level. All data are combined from at least 3 independent experiments, and mean values are shown.
In general, at the lipid-class level, our data from mouse correlate very well with known literature values derived from human platelets (Figure 3D; supplemental Table 2) yielding a Pearson coefficient of R2 = 0.9888. In individual cases, the concentrations can vary significantly. These can be derived from difference in the measurement strategy, dietary effects or enzyme activities (lecithin-cholesterol acyltransferase, etc.), which differ in genetic backgrounds and the absence of synchronized study conditions. Nevertheless, the majority of the reported concentrations at lipid class level in mouse and human platelets, such as those of PE, PS, PG, phosphatidic acid (PA), PC, PC P/O, SM, Cer, ChoE, lysophosphatidylcholine (LPC), FFA, and hexose Cer (HexCer), correlate very well, underscoring the value of the mouse model for disease-related lipidomics. Even if the total amount at the lipid classes is roughly conserved in mouse and man lipids are distinct and quite diverse with a sustained core of lipids at the lipid species (Figure 3E; supplemental Table 3). Compared with mouse data, human lipidome (489 identified lipids) displays slightly higher diversity with 58% shared lipids (Figure 3E; supplemental Table 3).
Lipids acutely generated during platelet activation
Most important pathways contributing to platelet activation include GPVI- and PAR-dependent signaling. Using collagen-related peptide (CRP) and thrombin separately, both pathways could be studied independently. Platelet activation is a very rapid and dynamic process that takes place within seconds to minutes.39 To study the platelet lipidome after activation, all experiments were conducted at a time point of 5 minutes, using the following standard stimuli (1) CRP, (2) thrombin, and (3) a combination of both. The 5-minute time point was chosen based on previous publications.19,38 Lipid precursors can be rapidly converted into downstream signaling lipids upon platelet activation.40
The majority of lipids was not altered during activation (Figure 4A; supplemental Figures 4-5). Irrespective of the chosen stimulus, the platelet lipidome remained highly stable with respect to the major membrane components, with a change in only 1 (PI 18:0-20:4) from the 15 most abundant lipids. This stability guarantees membrane integrity during activation. The majority of significantly regulated lipids were medium- to low-abundance lipids. Here, 74 lipids changed by a factor of 2 (P < .05) for thrombin (24), for CRP (46), and for thrombin plus CRP (54) (Figure 3B). Across all stimuli, the most prominent changes were observed for PA 18:1-20:4, which showed, on average, a 42-fold upregulation, and LM such as 12-HETE which showed a 48-fold increase. In addition to lipids that were regulated across all stimuli, a distinct set of lipids were identified for each individual stimulus (supplemental Figures 6-7; supplemental Table 4). CRP caused an upregulation of lyso-PL and a distinct population of LM compared with all other stimuli. Thrombin downregulated a high number of TAG, most likely for energy demands. Only combined and enhanced stimuli significantly upregulated all LM.
Absolute quantification of the activated platelet lipidome reveals arachidonic acid metabolism as the most dynamic network. (A) Regulated and nonregulated lipid species numbers across different stimuli. (B) Venn diagram of significantly regulated lipid species in wild-type platelets across 3 different types of activation (thrombin, CRP, thrombin + CRP). (C) Scatter plots displaying selected examples of pairs of lipids whose relative abundance is either positively (top left panel) or negatively (top right panel) correlated. Black lines indicate linear fit (bottom panel). Analysis of the fraction of correlations connecting lipids of the same lipid class (red) or different lipid classes (blue), as function of correlation strength. (D) Hierarchical clustering of the lipid-lipid correlation and anticorrelation matrix. Rows and columns correspond to the 384 quantified lipid species. Black boxes indicate clusters of strongly correlated and anticorrelated lipids. Lipid cluster numbers are indicated on the right. (E) Number of lipids in each cluster sorted by lipid class. (F) Network visualization of the lipid-lipid correlations. Edges are correlations of r ≥ 0.85. Nodes are lipids. Node size represents the degree of connectivity and the node color represents the analyzed lipid class (see inset). (G-I) The network during activation by thrombin, CRP, or thrombin + CRP; pure red indicates a fold ≥2 and pure green a fold change ≤0.5. Data are combined from 3 independent biological experiments and mean values are shown.
Absolute quantification of the activated platelet lipidome reveals arachidonic acid metabolism as the most dynamic network. (A) Regulated and nonregulated lipid species numbers across different stimuli. (B) Venn diagram of significantly regulated lipid species in wild-type platelets across 3 different types of activation (thrombin, CRP, thrombin + CRP). (C) Scatter plots displaying selected examples of pairs of lipids whose relative abundance is either positively (top left panel) or negatively (top right panel) correlated. Black lines indicate linear fit (bottom panel). Analysis of the fraction of correlations connecting lipids of the same lipid class (red) or different lipid classes (blue), as function of correlation strength. (D) Hierarchical clustering of the lipid-lipid correlation and anticorrelation matrix. Rows and columns correspond to the 384 quantified lipid species. Black boxes indicate clusters of strongly correlated and anticorrelated lipids. Lipid cluster numbers are indicated on the right. (E) Number of lipids in each cluster sorted by lipid class. (F) Network visualization of the lipid-lipid correlations. Edges are correlations of r ≥ 0.85. Nodes are lipids. Node size represents the degree of connectivity and the node color represents the analyzed lipid class (see inset). (G-I) The network during activation by thrombin, CRP, or thrombin + CRP; pure red indicates a fold ≥2 and pure green a fold change ≤0.5. Data are combined from 3 independent biological experiments and mean values are shown.
Significant solid regulation of numerous lipids was observed in lipid classes such as TAG, DAG, PA, PI, and different lyso-PL, and should not be neglected because the change expressed in absolute numbers is making up to 3000 molecules per cell (eg, PI 18:0-20:4) and will therefore strongly impact on the release of FFA during platelet activation.
Given the distinct changes observed for the 2 different stimuli, we next analyzed the coregulation of lipid concentrations at the individual lipid FA species level to elucidate higher organizational changes involved in platelet activation (Figure 4). Comparing 384 lipids across the different stimuli revealed pairs of positively and negatively correlated lipids (Figure 4C). For example, PA 18:0-20:4, PA 18:1-20:4, LPI 18:0 are positively correlated whereas PI 18:0-20:4 and PC 20:4-18:0 are negatively correlated with 12-HETE. Using the Pearson correlation computed for any pair of lipids, 12 distinct clusters of correlated and anticorrelated lipids (Pearson correlation ≥0.85) during platelet activation were revealed (Figure 4D,E).
PLs were distributed over almost all of the different clusters except C1, which was exclusively occupied by the SL and GL classes. Positive correlation within and above different classes indicates that lipids are derived from related pathways, whereas negative correlation indicates that breakdown products of 1 class are used for the synthesis of other lipid species. Indeed, this is the case for LM, PL, and GL species in C3-C6. The correlation reflects their metabolic proximities (Figure 4C). AA itself correlates positively with their downstream LM products during the combined stimulus, whereas it is rather stable under CRP or thrombin stimulation.
Lipid abundance, and the individual change of each species, is difficult to access from a hierarchical view, indicating that this is a suboptimal way of presenting coregulation within the lipidome. We therefore converted the lipid-lipid correlation matrix into a network. Here, nodes represent individual lipid species and edges represent correlations or anticorrelations of 0.85 or above. Cohesion across different clusters was elucidated by mapping different individual lipid features, such as fold change and lipid class onto the generated network (Figure 4F-I; supplemental Figure 8).
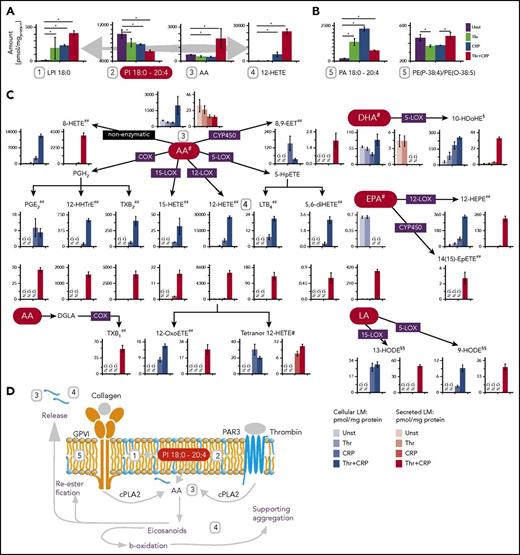
The input stimuli have a particularly strong effect on LM levels, where most of the mediators are produced and secreted in high amounts (Figure 5). A color-coded network with the fold change of the individual stimulus compared with the control revealed a common regulation direction of mediators across the stimuli used, and also revealed a distinct regulation of PL and GL upon platelet activation (Figure 4G-I). The absolute quantification suggests that, for all stimuli, PI 18:0-20:4 is the major precursor for the production of LPI, AA, and its downstream LM such as 12-HETE (Figure 5A). Off note, the next abundant and regulated AA-carrying PI is 100-fold less concentrated. Furthermore, many AA-carrying PA molecules (PA 18:1-20:4, PA 16:0-20:4, PA 18:0-20:4) are highly upregulated after thrombin and CRP treatment (Figure 5B). This is specific for AA-PAs, was not observed for other lipid classes such as PC, indicates reesterification of AA and suggests that AA-PAs may play an important role as an endogenous positive feedback signal to amplify PIP2-specific phospholipase C in mouse platelets.41,42 In general, thrombin stimulation had only a minor effect on LM level, whereas CRP alone was able to trigger the expression of AA-related LMs such as LTB4, TXB2, 12-HETE, 12-HHTrE, 8-HETE, tetranor 12-HETE, and 12-OxoETE (Figure 5C) and only a combined stimulus the secretion of all mediators. The combined stimulus that is the closest to in vivo conditions specifically elevated secreted mediators such as PGE2, EPA, 14(15)-EpETE, 8,9-EET, and LTB4, indicating that within this chosen short time frame (5 minutes) both stimuli are needed for a proper response to these mediators. To rule out that the higher concentration of stimulus is responsible for mediator release, the activation was further conducted with individual higher CRP and Thr concentrations. This confirms that EPA, 14(15)-EpETE, 8,9-EET, and LTB4 are only secreted by Thr/CRP stimulus (supplemental Figure 9), which may indicate that only a stimuli cross-talk is able to trigger LM key players simultaneously (Figures 4I and 5A). In summary, PI 18:0-20:4 is the major precursor for AA and for most of the produced mediators in mouse and human (supplemental Figure 10). CRP and the combined stimulus induce a stronger mediator response than thrombin alone. AA is reesterified, and many mediators are released from platelets (Figure 5D). Thus, the total number of released mediators and their concentrations are similar to or higher than those found inside platelets, underscoring a similar importance for intracellular and extracellular signaling. Off note, the individual identified mediators in activated mouse and human platelets slightly differ in occurrence and concentration but the overall trends are conserved over different stimulations (supplemental Figure 10).
Mediator production and release in platelets is stimulus dependent and centered. (A) Concentration (pmol/mgprotein) of 12-HETE, arachidonic acid and PI 18:0-20:4. (B) Bar graphs of various mediator precursors during different stimuli. (C) Analyzed mediators in platelets and their corresponding secretome level. Bar graphs with black-framed colors display mediators in platelets; bars with red-framed colors indicate mediators in the corresponding secretome. (D) Measured lipid regulation during platelet activation. The absolute quantities are reported in pmol/mgprotein. Data were combined from 3 independent experiments, displayed as mean values, and the error is represented as standard deviation of the mean. #The lipid class of free FAs (FFA); ##Eicosanoids; §Docosanoids; §§Octadecanoids. N.D., not detectable.
Mediator production and release in platelets is stimulus dependent and centered. (A) Concentration (pmol/mgprotein) of 12-HETE, arachidonic acid and PI 18:0-20:4. (B) Bar graphs of various mediator precursors during different stimuli. (C) Analyzed mediators in platelets and their corresponding secretome level. Bar graphs with black-framed colors display mediators in platelets; bars with red-framed colors indicate mediators in the corresponding secretome. (D) Measured lipid regulation during platelet activation. The absolute quantities are reported in pmol/mgprotein. Data were combined from 3 independent experiments, displayed as mean values, and the error is represented as standard deviation of the mean. #The lipid class of free FAs (FFA); ##Eicosanoids; §Docosanoids; §§Octadecanoids. N.D., not detectable.
Impact of Smpd1 on the platelet lipidome
To examine the applicability of the developed quantitative platelet lipidomics analysis in a disease model, we analyzed the lipidome of platelets from gene-targeted mice lacking Smpd1 (Smpd1−/−) to characterize the functional role of SMPD1 in platelet lipid signaling and to clarify which lipids contribute to defective secretion in Smpd1−/− platelets. For that purpose, platelets were isolated from Smpd1−/− mice and their wild-type littermates (Smpd1+/+). Compared with other storage diseases, the lipidome of platelets from Smpd1−/− mice is stable, and <20% of the quantified lipids were affected by the genetic SMPD1 knockout (Figure 6A; supplemental Figures 11-12). By applying the agonists CRP and thrombin, we expected a strong remodeling of the platelet sphingolipidome, and that the SM to Cer ratio would move toward a reduced Cer level. This, however, was only observed for Cer with the long-chain base (LCB) d18:2 (supplemental Figures 11 and 13). SPC was highly upregulated (by a factor of 10) in SMPD1-deficient platelets to give a final concentration of 20 µM (Figure 6A-B), which is 800-fold higher than the concentration found in human plasma (25 nM).43 Other SL lipids, such as So (sphingosine) and Sa (sphinganine), were simultaneously decreased, indicating a shift in SL metabolism toward SPC (Figure 6B-C).
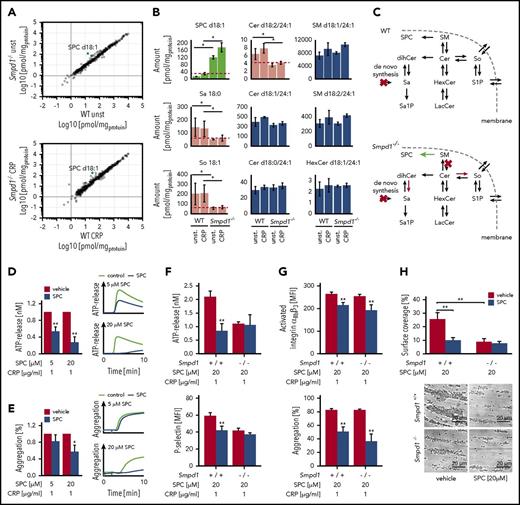
Quantitative lipidomics reveals SPC dysregulation at the trans-Golgi network in the Smpd1−/−mouse strain. (A) Correlation of murine lipid concentrations between wild-type (WT) and acidic sphingomyelinase knockout (Smpd1−/−) mutant mice in resting platelets (top panel) and after stimulation with CRP (1 µg/mL; bottom panel). (B) Regulated and nonregulated SLs in resting and activated platelets. Data from 3 independent experiments were combined. (C) SL metabolism in WT and Smpd1−/− mice indicates a shift toward lysosphingomyelin. (D) ATP release in murine wild-type platelets pretreated with SPC or vehicle control for 2 minutes followed by a stimulation with CRP. Arithmetic means ± SEM (n = 6; left side) and characteristic graphs (right side) are shown. (E) Platelet light transmission aggregometry in murine wild-type platelets pretreated with SPC or vehicle control followed by a stimulation with CRP. Arithmetic means ± SEM (n = 6; left side) and characteristic graphs (right side) are shown. (F) Platelet secretion of dense (ATP release) and α (P-selectin) granules in Smpd1+/+ and Smpd1−/− platelets in the presence of SPC (20 µM) or vehicle control. (G) Platelet integrin αIIbβ3 activation (top panel) and aggregation (bottom panel) of Smpd1+/+ and Smpd1−/− platelets in the presence of SPC or vehicle control following stimulation with CRP. (H) Arithmetic means ± SEM (n = 6; top panel) and representative phase-contrast images (bottom panel) of platelet surface coverage after perfusion from blood from Smpd1−/− and wild-type mice over a collagen-coated surface (200 µg/mL) for 5 minutes in the presence (gray bars) or absence (black bars) of SPC and at a shear rate of 1700−sec. Scale bar equals 20 µm. *(P < .05) and **(P < .01) indicate statistically significant differences. LacCer, lactosylceramide; S1P, sphingosine-1-phosphate; Sa, sphinganine; Sa1P, sphinganine-1-phosphate; SEM, standard deviation of the mean; So, sphingosine.
Quantitative lipidomics reveals SPC dysregulation at the trans-Golgi network in the Smpd1−/−mouse strain. (A) Correlation of murine lipid concentrations between wild-type (WT) and acidic sphingomyelinase knockout (Smpd1−/−) mutant mice in resting platelets (top panel) and after stimulation with CRP (1 µg/mL; bottom panel). (B) Regulated and nonregulated SLs in resting and activated platelets. Data from 3 independent experiments were combined. (C) SL metabolism in WT and Smpd1−/− mice indicates a shift toward lysosphingomyelin. (D) ATP release in murine wild-type platelets pretreated with SPC or vehicle control for 2 minutes followed by a stimulation with CRP. Arithmetic means ± SEM (n = 6; left side) and characteristic graphs (right side) are shown. (E) Platelet light transmission aggregometry in murine wild-type platelets pretreated with SPC or vehicle control followed by a stimulation with CRP. Arithmetic means ± SEM (n = 6; left side) and characteristic graphs (right side) are shown. (F) Platelet secretion of dense (ATP release) and α (P-selectin) granules in Smpd1+/+ and Smpd1−/− platelets in the presence of SPC (20 µM) or vehicle control. (G) Platelet integrin αIIbβ3 activation (top panel) and aggregation (bottom panel) of Smpd1+/+ and Smpd1−/− platelets in the presence of SPC or vehicle control following stimulation with CRP. (H) Arithmetic means ± SEM (n = 6; top panel) and representative phase-contrast images (bottom panel) of platelet surface coverage after perfusion from blood from Smpd1−/− and wild-type mice over a collagen-coated surface (200 µg/mL) for 5 minutes in the presence (gray bars) or absence (black bars) of SPC and at a shear rate of 1700−sec. Scale bar equals 20 µm. *(P < .05) and **(P < .01) indicate statistically significant differences. LacCer, lactosylceramide; S1P, sphingosine-1-phosphate; Sa, sphinganine; Sa1P, sphinganine-1-phosphate; SEM, standard deviation of the mean; So, sphingosine.
To elucidate the role of SPC in platelet function, we first conducted adenosine triphosphate (ATP) release and aggregation measurements in wild-type platelets in the presence or absence of exogenous SPC. Platelets were pretreated for 2 minutes with increasing concentrations of SPC or vehicle control, respectively, and activation-dependent ATP release and aggregation were estimated after stimulation with 1 µg/mL CRP (Figure 6D). Pretreatment with low (5 µM) and high doses (20 µM) of SPC significantly decreased platelet-dense granule secretion compared with vehicle-treated platelets. Moreover, CRP-induced platelet aggregation was significantly impaired after pretreatment with high doses of SPC. To elucidate whether SPC affects platelet activation in a SMPD1-dependent manner, we performed further studies in Smpd1−/− platelets. As illustrated in Figure 6F, SPC significantly reduced dense (reflected by ATP release) and α (reflected by P-selectin exposure) granule secretion as compared with the levels found in Smpd1−/− platelets and had no additive effect in SMPD1-deficient platelets. However, integrin αIIbβ3 activation and platelet aggregation were significantly affected independently of SMPD1 and only by high doses of SPC (Figure 6E,G). However, in vitro thrombus formation on collagen-coated surfaces under high arterial shear rates was significantly reduced in Smpd1+/+ platelets by addition of SPC to a comparable level found in Smpd1−/− platelets. In contrast, treatment of Smpd1−/− platelets even with high-dose SPC had no effect on thrombus formation under high shear rates pointing to a pivotal and SMPD1-dependent role for SPC in the regulation of arterial thrombus formation.
Discussion
This study provides the first quantitative investigation of the mouse platelet lipidome and a validation of major finding in human platelets. Combining structural with quantitative results was the key to discover which species contribute most to the polyunsaturated FA (PUFA) arachidonic acid (20:4, AA) centered platelet lipidome. In platelets, AA occurs more often than in other lipidomes such as erythrocytes, plasma, or heart tissue, where the more highly saturated FA (palmitic [16:0], stearic [18:0], and oleic acid [18:1]) are predominantly esterified to phospholipid classes (PL).44 The use of PUFA as central lipidomic building blocks (>20 mol %) enhances stability of the ordered lipid-raft domains, due to an increased order difference between them and the coexisting nonraft domains,45 which could stabilize signaling events in platelets. In addition, these PUFA-PL serve as precursors for the production of mediators such as prostaglandins, which orchestrate in a positive and negative feedback-loop platelet signaling after stimulation.46 Applying the developed assay across different stimuli was decisive to identify PI 20:4-18:0 as the major precursor for the production of AA and its downstream mediators in mice and humans. However, based on our results, it is also clear that PI is not the only source contributing to the total amount of LMs produced upon platelet activation. The massive burst of mediators and lysolipids has several important implications for platelet physiology: (1) an enormous quantity of signaling molecules are released, triggering paracrine and autocrine downstream targets, such as IP receptor and TXA2 receptor.47,48 (2) PI-derived mediators also serve as an energy source for the high-energy demand during platelet aggregation, or are reesterified for later use.19 (3) The drop in PI 20:4-18:0 and the increase in lyso-PI 18:0 potentially have a profound effect on membrane fluidity and curvature, supporting granule release and platelet aggregation.45,49
CRP and thrombin induced no major lipidomic shifts after platelet activation, and the average overall dynamics was consistently <20% in all quantified lipids, indicating distinct triggering of pathway-dependent events and a stable lipidome. Of note, the platelet lipidome becomes modified under several different conditions as shown for chronic vascular inflammation/atherosclerosis in patients with coronary artery disease or myocardial infarction.50 Lipids derived from activated platelets are important mediators of vascular inflammation. In particular, specific platelet-derived lipids are recruited into microparticles that are produced following platelet activation51 and may trigger proinflammatory responses via intercellular interactions.
To validate our method in a disease model with genetic deficiency in lipid signaling, where platelet function and thrombus formation is disturbed,6 and to demonstrate that the analysis is sensitive enough to monitor disease-related lipidomic changes, we investigated activation-dependent lipidome modifications in platelets from Smpd1-deficient mice.52 The absence of a major enzyme of the SL metabolism resulted in a still stable platelet lipidome. In contrast, the SL metabolism itself shifted toward enhanced production of SPC while the sphingosine and sphinganine levels were reduced, which in general indicates that the cis to trans-Golgi system, where these lipids are synthesized, is disturbed. Finally, this impaired cis to trans-Golgi system could be manifested, especially in modified platelet granule trafficking and biogenesis because the trans-Golgi network in megakaryocytes provides a central platform for the development of α-, δ-granules as well as lysosomes during megakaryopoiesis/thrombopoiesis.53,54 In Smpd1−/− platelets, the Cer generation using the sphingomyelinase pathway is perturbed without affecting platelet count,6 but with decreased platelet secretion and thrombus formation.6 Remarkably, this phenotype could be mimicked in wild-type platelets by administering SPC at physiological conditions, which led to impaired GPVI-dependent platelet activation. Treating Smpd1+/+ platelets with SPC leads to decreased in vitro thrombus formation, comparable to that in Smpd1−/− platelets, thus pointing to a direct correlation between SPC levels and observed platelet dysfunction in Smpd1−/− mice. This hypothesis was confirmed for platelet secretion, as there was no additional effect of genetic SMDP1 knockout in the absence or presence of high-dose SPC as compared with SPC-treated Smpd1+/+ samples, indicating a direct regulation of SPC levels and SPC-driven effects in platelet degranulation and thrombus formation by SMPD1. The inhibitory and mimicking effect of SPC on platelet function highlights the relevance of this lipid species for platelet granule secretion and thrombus formation. But remarkably, high-dose SPC significantly reduced platelet integrin αIIbβ3 activation and aggregation in Smpd1−/− platelets. We therefore speculate that high-dose SPC may affect platelet integrin activation and aggregation independently of Smpd1, for example, via activation of protein kinase A and modulation of vasodilator-stimulated phosphoprotein phosphorylation or via suppression of intracellular Ca2+ release.25 Smpd1−/− platelets did not alter Ca2+ signaling after activation.6 Thus, further studies will be necessary to dissect the underlying molecular mechanisms. In cardiomyocytes, SPC induces lipid raft formation and activates the PTEN/Akt1/mTOR pathway.55 In lung cancer cells, SPC activates Akt1.56 Akt1 and lipid rafts are both important signaling events in platelet activation and thrombosis.1,57,58
In general, minor plasma SLs such as not only sphingosine, sphinganine but also SPC modulate clotting factor activities potentially affecting thrombosis risk and contributing to the progression of thromboinflammatory and thrombo-occlusive diseases such as atherosclerosis, myocardial infarction. and ischemic stroke.59 The present study identified SPC as a novel regulator of platelet degranulation and subsequent thrombus formation. SPC also fosters proinflammatory events in cerebral arteries,58,60 contributing to progression of atherosclerosis and cardiovascular events. High plasma levels of SPC are associated with metabolic syndrome,43 which is still 1 major risk factor for cardiovascular diseases and thrombo-occlusive disorders. Thus, SPC could be useful as antiplatelet therapy in thrombo-occlusive diseases.
The present study is the first to quantify the platelet lipidome, which comprises almost 400 lipid species and covers a concentration range of 7 orders of magnitude. The platform presented here uniquely permits a systematic assessment of the lipidomics network in resting and activated murine platelets, and demonstrates the feasibility of performing absolute and comprehensive quantitative platelet-lipidome analysis. The operation of our platform yielded in identification of SMPD1 as a specific modulator of the platelet lipidome, involved in the regulation of SPC levels being critical to platelet-dense granule release and thrombus formation.
This approach opens new doors to study the functional consequence of the (genetic) deficiency of particular proteins and other molecules involved in platelet lipid metabolism regulation. Thus, the application of this workflow might be helpful to identify lipids affected in (patho-) physiological platelet activation and modified in thromboinflammatory diseases such as atherosclerosis, coronary artery disease, or acute myocardial infarction with the potential to identify novel biomarkers or targets for antithrombotic treatment strategies.
The full-text version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin, the Bundesministerium für Bildung, and by grants from the Deutsche Forschungsgemeinschaft (DFG) (CRC 240 [R.A., O.B., A.S., and M.G.]; BO 3786/1-1 [O.B.], KFO 274 [O.B. and M.G.]), the fortüne research programme, the German Cardiac Foundation, and the Dr Karl Kuhn Foundation. We received further support from the Federal Ministry of Education and Research (BMBF) in the German Network Bioinformatics Infrastructure (de.NBI) initiative grants No 031L0108A and 031 A 534B.
Authorship
Contribution: B.P., S.G., C.C., P.M., D.K., M.-C.M., F.L., O.B., and R.A. designed and performed experiments; B.P., S.G., C.C., P.M., D.K., C.H., N.H., F.L., O.B., and R.A. analyzed and interpreted data; A.S. and M.G. discussed the content; and O.B., B.P., and R.A. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Robert Ahrends, Leibniz-Institut für Analytische Wissenschaften -ISAS -e.V., Otto-Hahn-Str.6b, 44227 Dortmund, Germany; e-mail: robert.ahrends@isas.de; or Oliver Borst, Department of Cardiology and Cardiovascular Medicine, University of Tübingen, Tübingen, Germany; e-mail: oliver.borst@med.uni-tuebingen.de.
REFERENCES
Author notes
O.B. and R.A. contributed equally and share last authorship.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal