Key Points
Mutations in a VHL cryptic exon may be found in patients with familial erythrocytosis or VHL disease.
Synonymous mutations in VHL exon 2 may induce exon skipping and cause familial erythrocytosis or VHL disease.
Abstract
Chuvash polycythemia is an autosomal recessive form of erythrocytosis associated with a homozygous p.Arg200Trp mutation in the von Hippel-Lindau (VHL) gene. Since this discovery, additional VHL mutations have been identified in patients with congenital erythrocytosis, in a homozygous or compound-heterozygous state. VHL is a major tumor suppressor gene, mutations in which were first described in patients presenting with VHL disease, which is characterized by the development of highly vascularized tumors. Here, we identify a new VHL cryptic exon (termed E1′) deep in intron 1 that is naturally expressed in many tissues. More importantly, we identify mutations in E1′ in 7 families with erythrocytosis (1 homozygous case and 6 compound-heterozygous cases with a mutation in E1′ in addition to a mutation in VHL coding sequences) and in 1 large family with typical VHL disease but without any alteration in the other VHL exons. In this study, we show that the mutations induced a dysregulation of VHL splicing with excessive retention of E1′ and were associated with a downregulation of VHL protein expression. In addition, we demonstrate a pathogenic role for synonymous mutations in VHL exon 2 that altered splicing through E2-skipping in 5 families with erythrocytosis or VHL disease. In all the studied cases, the mutations differentially affected splicing, correlating with phenotype severity. This study demonstrates that cryptic exon retention and exon skipping are new VHL alterations and reveals a novel complex splicing regulation of the VHL gene. These findings open new avenues for diagnosis and research regarding the VHL-related hypoxia-signaling pathway.
Introduction
Congenital erythrocytosis represents a heterogeneous group of rare disorders. Genetic changes affecting all parts of the regulatory pathway of erythropoiesis, including oxygen sensing, erythropoietin sensitivity, and hemoglobin oxygen affinity, have been described in patients with congenital erythrocytosis. The detection of underlying genetic changes in patients with presumed hematological pathology may have important implications for adequate clinical management. However, even with the use of next-generation sequencing panel diagnostics, the underlying genetic cause of presumed congenital erythrocytosis has been identified in fewer than one third of patients in most published cohorts.
The molecular basis of von Hippel-Lindau (VHL)–related congenital erythrocytosis was first described in the autonomous Russian Republic of Chuvashia, where this condition is an endemic disorder.1 Chuvash polycythemia is frequently associated with rubor, vertebral hemangiomas, varicose veins, and low blood pressure. Chuvash patients have reduced survival rates associated with a higher prevalence of arterial and venous thromboses and pulmonary hypertension in addition to hemorrhagic events.2
Chuvash polycythemia arises from a homozygous c.598C>T p.Arg200Trp (R200W) mutation in the VHL gene. This specific VHL R200W mutation has also been identified in combination with other VHL mutations (compound heterozygosity), and other missense VHL mutations in both alleles have been described in patients with congenital erythrocytosis.3,4 Interestingly, it has been described in some unexplained cases of patients with erythrocytosis carrying only one heterozygous mutation.4-6
VHL is located on 3p25-26 and has been reported to contain 3 exons (E1, E2, and E3). The commonly described VHL transcript contains the 3 spliced exons that encode a 213 amino acid (aa) protein (pVHL213; also termed pVHL30) and a smaller isoform (pVHL160 or pVHL19) initiated from an in-frame internal translation start site.7 A naturally occurring splice variant, expressed at low levels in some tissues, comprises E1 directly spliced to E3 and is translated into a protein product termed pVHL172 (pVHLΔE2), the functions of which are still under investigation.8-12 pVHL213 and pVHL160 are involved in a variety of functions, the most studied being the regulation of the cellular oxygen-sensing pathway. The main player of this pathway is the hypoxia inducible factor (HIF). Under normal oxygen supply, the α subunits of HIF (HIF-1α, -2α, and -3α) are hydroxylated by the prolyl-4 hydroxylase domain enzymes (PHD1, -2, and -3) and subsequently targeted by pVHL, a subunit of an E3 ubiquitin-ligase complex that promotes HIF-α ubiquitination and subsequent proteasomal degradation.13,14 Under hypoxic conditions or when VHL is mutated, HIF-α remains stable and heterodimerizes with HIF-β, constituting a functional HIF. HIF transcriptionally activates a variety of genes involved in adaptation to reduced oxygen supply (eg, erythropoiesis, angiogenesis, metabolism, and cell survival). Dysregulation of the hypoxia pathway13,14 is central to the development of erythrocytosis1,4 (via upregulation of erythropoietin, a HIF-2α target gene), but also in the development of tumors.15 Indeed, VHL is a tumor suppressor gene, heterozygous mutations of which are associated with VHL disease (Figure 1A).16,17 VHL disease, described in 1936, is an autosomal dominant disorder with high penetrance characterized by the development of highly vascularized tumors like central nervous system and retinal hemangioblastomas, pancreatic neuroendocrine tumors, pheochromocytomas, and clear cell renal cell carcinomas.18-20
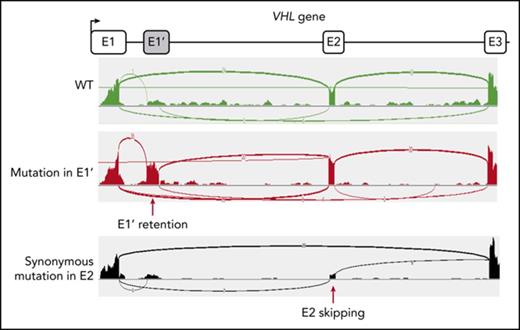
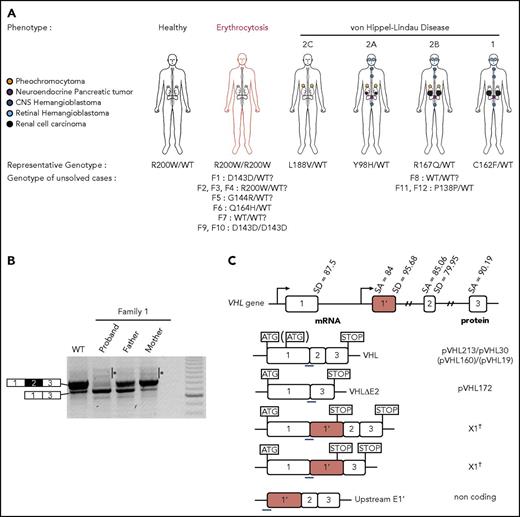
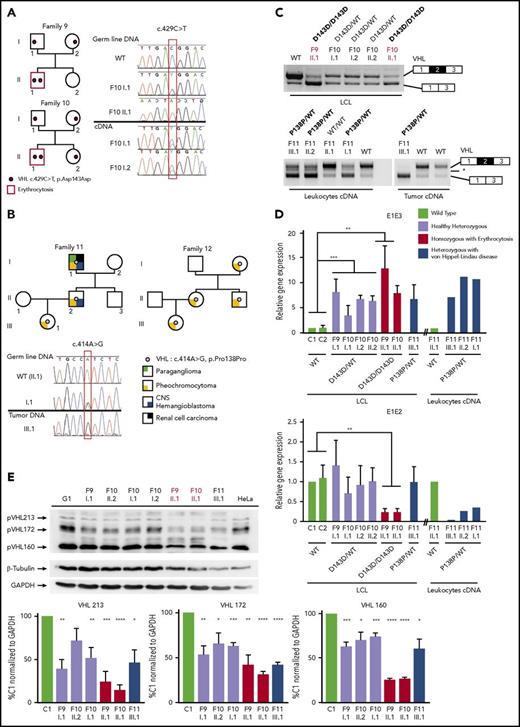
Clinical manifestations of patients carrying VHL mutations and identification of a new VHL spliced isoform containing a cryptic exon. (A) Mutations in the VHL gene predispose to different phenotypes. VHL disease is characterized by the development of central nervous system (CNS) and retinal hemangioblastomas, neuroendocrine pancreatic tumors, pheochromocytomas, and clear cell renal cell carcinomas. Chuvash polycythemia (erythrocytosis) is characterized by elevated red blood cell numbers. This study describes families with typical VHL-related phenotypes associated with an unexpected VHL status (ie, either synonymous mutations or no alterations in VHL). (B) Reverse transcription polymerase chain reaction (RT-PCR) using primers specific for E1 and E3 was performed on messenger RNA (mRNA) extracted from lymphoblastoid cell lines (LCLs) established from controls and patients of family 1 (F1). (C) Schematic representation of the VHL gene and its products. The different VHL exons are represented on a scale: E1, 340 bp from the ATG initiation codon; E1′, 259 bp; E2, 123 bp; E3, 179 bp to the Stop termination codon. The full-length VHL mRNA isoform encodes pVHL213 (also named pVHL30). VHL E1 contains an internal translation initiation codon that initiates the production of pVHL160 protein (pVHL19). The isoform containing E1 spliced in phase with E3 encodes pVHL172 that lacks E2. The isoforms containing E1 spliced with E1′ may theoretically encode a protein termed X1 of 193 aa (114 aa encoded by E1, and 79 aa encoded by E1′). Consensus values of donor (SD) and acceptor (SA) splice site sequences are indicated above the VHL gene, as calculated by the human splicing finder in silico tool. Horizontal blue lines indicate the location of probes used in TaqMan assays. *Denotes larger fragments that were subsequently cloned and sequenced. †The isoforms containing E1′ spliced with VHL exons were identified by cloning and sequencing in the laboratory but were described later by the National Center for Biotechnology Information as transcripts able to produce a protein termed X1. WT, wild type.
Clinical manifestations of patients carrying VHL mutations and identification of a new VHL spliced isoform containing a cryptic exon. (A) Mutations in the VHL gene predispose to different phenotypes. VHL disease is characterized by the development of central nervous system (CNS) and retinal hemangioblastomas, neuroendocrine pancreatic tumors, pheochromocytomas, and clear cell renal cell carcinomas. Chuvash polycythemia (erythrocytosis) is characterized by elevated red blood cell numbers. This study describes families with typical VHL-related phenotypes associated with an unexpected VHL status (ie, either synonymous mutations or no alterations in VHL). (B) Reverse transcription polymerase chain reaction (RT-PCR) using primers specific for E1 and E3 was performed on messenger RNA (mRNA) extracted from lymphoblastoid cell lines (LCLs) established from controls and patients of family 1 (F1). (C) Schematic representation of the VHL gene and its products. The different VHL exons are represented on a scale: E1, 340 bp from the ATG initiation codon; E1′, 259 bp; E2, 123 bp; E3, 179 bp to the Stop termination codon. The full-length VHL mRNA isoform encodes pVHL213 (also named pVHL30). VHL E1 contains an internal translation initiation codon that initiates the production of pVHL160 protein (pVHL19). The isoform containing E1 spliced in phase with E3 encodes pVHL172 that lacks E2. The isoforms containing E1 spliced with E1′ may theoretically encode a protein termed X1 of 193 aa (114 aa encoded by E1, and 79 aa encoded by E1′). Consensus values of donor (SD) and acceptor (SA) splice site sequences are indicated above the VHL gene, as calculated by the human splicing finder in silico tool. Horizontal blue lines indicate the location of probes used in TaqMan assays. *Denotes larger fragments that were subsequently cloned and sequenced. †The isoforms containing E1′ spliced with VHL exons were identified by cloning and sequencing in the laboratory but were described later by the National Center for Biotechnology Information as transcripts able to produce a protein termed X1. WT, wild type.
In patients carrying VHL mutations, the precise mechanistic aspects that underpin the different phenotypes remain obscure. Although most patients carry mutations in the VHL gene that induce a partial or complete loss of protein function, some cases remain unsolved. Indeed, some patients with erythrocytosis have been found to be heterozygous rather than homozygous for the expected alteration4-6 or carry homozygous synonymous mutations that leave the amino acid sequence intact. In addition, some patients present with VHL disease in the absence of identified mutations or deletions in VHL or carry heterozygous synonymous mutations. Here, we report an investigation of 12 families linked to unexplained disease, including 9 families with erythrocytosis and 3 with VHL disease (Figure 1A). This study led to the discovery of a novel cryptic exon in the VHL gene and complex regulation of VHL splicing.
Methods
Complete materials and methods are detailed in the supplemental Material and methods (available on the Blood Web site).
Study approval
Informed consent for medical diagnosis and research was obtained from the patients and their relatives. This study was agreed by the Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale (French ethical committee) Paris-Sud at Bicêtre Hospital.
Sanger sequencing
Exons and exon-intron junctions of the VHL gene were sequenced from DNA extracted from whole blood, as previously described.21
WGS
Whole-genome sequencing (WGS; for families F2, F3, and F7) was performed at the clinically accredited Molecular Diagnostics Laboratory at the John Radcliffe Hospital using the Hi-Sequation 4000 platform (Illumina Inc., San Diego, CA) in high-throughput mode.22,23 Analysis of single-nucleotide variants, short insertions/deletions, and copy number variants was conducted and is explained in detail in the supplemental Methods.
Transcript detection and quantification
After reverse-transcription reactions (ThermoScientific), exon-specific polymerase chain reaction (PCR) was performed using primers localized to flanking E1 and E3 exons. TaqMan real-time PCR were performed on 20 ng of complementary DNA (cDNA) with the quantitative PCR Mastermix (Eurogentec). Quantification of RPLP0 transcripts was used as internal control. The thresholds were determined using dilutions of plasmids containing coding sequences of each gene.
RNA sequencing
Library construction was performed with SureSelect Strand-Specific RNA Library Prep for the Illumina Multiplexed kit (Agilent Technologies). After purification (Macherey-Nagel), the fragment size of libraries was controlled using the 2200 TapeStation system (Agilent Technologies). Ten pM of each library was pooled and prepared according to the denaturing and diluting library protocol for Hiseq and GAIIx (Illumina) for cluster generation on the cBot system. Paired-end sequencing was carried out in a single lane on the HiSeq 2500 system (Illumina). Processing of reads is detailed in the supplemental Methods.
Reporter assays
The plasmid encoding the WT hemagglutinin-tagged VHL was kindly provided by William G. Kaelin Jr. An expression plasmid for the hypothetical X1 protein was constructed by subcloning the coding sequence after its synthesis (Life Technologies). Mutations of interest were introduced by site-directed mutagenesis (New England Biolabs); 786-O cells were transfected (Ozyme) with constructs encoding WT or mutant proteins, firefly luciferase under the control of hypoxia response elements, and Renilla luciferase for normalization.24 After 24 hours, luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega).
Western blotting
Cell lysates from the luciferase reporter assay were loaded into a Bis-Tris Mini Gel (4% to 12%; Invitrogen). After transfer, the membrane (GE Healthcare) was subsequently incubated with a mouse anti-hemagglutinin antibody (BioLegend) and then a goat anti-mouse horseradish peroxidase–conjugated antibody (Jackson Immuno Research). Western blot using the mouse monoclonal antibody JD-1956 (patent #14305925.1-1402-2014; Centre National de la Recherche Scientifique Etablissement Français du Sang) raised against human VHL was performed as described.8
Minigene experiments
Minigene constructs were prepared in pCas2 plasmid, containing 2 artificial exons A and B (as described by Gaildrat et al25 ), between which VHL E1′ or VHL E2 with intronic flanking sequences were cloned. Cells were transfected (Ozyme) or nucleofected (Lonza). RNA was extracted 24 hours after transfection and reverse transcribed. PCR amplification was performed using the PCR GoTaqQ2 kit (Promega) with primers against artificial (A and B) exons. PCR products were resolved in a 2% agarose gel.
Results
Identification of new VHL spliced isoforms containing a cryptic exon
We first focused our study on a patient with erythrocytosis in whom a synonymous VHL c.429C>T p.Asp143Asp (D143D) mutation in the heterozygous state had previously been identified (F1; Table 1). No other mutations in the 3 VHL canonical exons had been identified in genomic DNA of this patient. RT-PCR using primers in E1 and E3 was performed using mRNA samples extracted from LCLs established from different family members. The results showed a strong decrease of the E1E2E3 isoform and an upregulation of the E1E3 isoform (Figure 1B) compared with WT LCLs. Some minor extra fragments of larger size were observed in this patient and his mother’s sample. Subcloning and sequencing of these fragments allowed us to identify new VHL transcripts that contained intronic sequences. This intronic sequence, which we termed the E1′ cryptic exon, was spliced to E1 at its 5′ end and to either E2 or E3 at its 3′ terminus (Figure 1C). We show that these isoforms were expressed in a variety of tissues and cell lines (supplemental Figure 1). Their translation may theoretically lead to the production of a protein of 193 aa that contains the first 114 aa encoded by E126 and additional 79 aa of unknown function encoded by E1′. During the course of our study, data from an automated computational analysis for an isoform containing the E1′ cryptic exon (E1E1′E2 isoform) were deposited in the National Center for Biotechnology Information. This isoform was predicted to encode a protein of 193 aa, named X1 (XP_011532380.1), with the same sequence (114 aa from E1 in addition to 79 aa encoded by E1′) (Figure 1C). The analysis of this region revealed strong conservation in primates, but a moderate to low conservation in more distant species (supplemental Figure 2A-C); notably, the splice sites were identical to canonical sites (ttcag/TC, AG/gtaag) and were highly conserved. In silico analysis of the donor and acceptor splice sites of E1′ showed similar consensus values compared with other VHL exons (Figure 1C, upper panel). The capacity to translate a potential X1 protein was only conserved in higher primates (supplemental Figure 2D). The deposited sequence was removed and replaced with a noncoding isoform containing E1′ spliced with VHL E2 and E3 (ENST00000477538.1; Figure 1C, isoform on the bottom). This isoform may be initiated by an alternative promoter. Indeed, the sequence located at the 5′ end of E1′ represents a transcriptionally active region, as illustrated by epigenetic marks (supplemental Figure 3). We confirmed the expression of an additional transcript initiated from the upstream region of E1′ (that we termed upstream E1′) in different tissues and cell lines (supplemental Figure 1).
Description of variants identified in VHL E1′ and VHL E2 with associated clinical manifestations
| Family no., patient . | Nucleotide variant on allele 1/allele 2 . | Impact on protein . | Year of birth/age at diagnosis . | Sex . | Hb, g/dL . | Ht, % . | Red cells, ×109/L . | EPO, mU/mL . | Phenotype . | Other mutation . |
|---|---|---|---|---|---|---|---|---|---|---|
| F1, II.1 | c.429C>T/c.340+770T>C | VHL p.Asp143Asp/X1 p.Ser179Pro? | 1991/15 y | M | 16.6 | 57 | 6.9 | 163 | Erythrocytosis, kidney ischemic infarct | — |
| F2, II.1 | c.598C>T/c.340+770T>C | VHL p.Arg200Trp/X1 p.Ser179Pro? | 1957/21 y | M | 19.9 | 67 | 22.2 | Erythrocytosis | — | |
| F3, II.1 | c.598C>T/c.340+770T>C | VHL p.Arg200Trp/X1 p.Ser179Pro? | 2003/4 mo | M | 17.9 | 54 | 7.5 | 60 | Erythrocytosis | — |
| F3, II.2 | c.598C>T/c.340+770T>C | VHL p.Arg200Trp/X1 p.Ser179Pro? | 2005/6 mo | M | 14.6 | 45 | Erythrocytosis | — | ||
| F4, II.1 | c.598C>T/c.340+694_711dup | VHL p.Arg200Trp/X1 p.Trp159X? | 1990/7 y | M | 20.6 | 64 | 8.25 | 49.9 | Erythrocytosis, deep vein thrombosis, intracerebral hemorrhage | FV Leiden |
| F5, II.2 | c.430G>A/c.340+694_711dup | VHL p.Gly144Arg/X1 p.Trp159X? | 1975/13 y | M | 16.2 | 60 | 5.9 | Erythrocytosis | — | |
| F6, II.1 | c.492G>T/c.340+574A>T* | VHL p.Gln164His/SA | 2014/6 mo | M | 15.6 | 49 | 6.9 | 1167 | Splenomegaly | — |
| F7 | c.340+816A>C/c.340+816A>C | X1 p.*194Serext*24/X1 p.*194Serext*24 | 7 y | M | 20 | 64 | 8.09 | 33 | Erythrocytosis | — |
| F8 | c.340+617C>G + c.340+648T>C†/WT | X1 p.Leu128Val+ X1 p.Leu138Pro/WT | N | N | N | N | VHL disease | — | ||
| F9, II.1 | c.429C>T/c.429C>T | VHL p.Asp143Asp/VHL p.Asp143Asp | 1997/5 mo | M | 16.2 | 49.3 | 5.2 | 186 | Erythrocytosis | Hb sitia |
| F10, II.1 | c.429C>T/c.429C>T | VHL p.Asp143Asp/VHL p.Asp143Asp | 2002/4.5 y | F | 22.5 | 64-77.4 | 12.6 | 264 | Erythrocytosis, splenomegaly | β thalassemia |
| F11 | c.414A>G/WT | VHL p.Pro138Pro/WT | N | N | N | N | VHL disease | — | ||
| F12 | c.414A>G/WT | VHL p.Pro138Pro/WT | N | N | N | N | VHL disease | — |
| Family no., patient . | Nucleotide variant on allele 1/allele 2 . | Impact on protein . | Year of birth/age at diagnosis . | Sex . | Hb, g/dL . | Ht, % . | Red cells, ×109/L . | EPO, mU/mL . | Phenotype . | Other mutation . |
|---|---|---|---|---|---|---|---|---|---|---|
| F1, II.1 | c.429C>T/c.340+770T>C | VHL p.Asp143Asp/X1 p.Ser179Pro? | 1991/15 y | M | 16.6 | 57 | 6.9 | 163 | Erythrocytosis, kidney ischemic infarct | — |
| F2, II.1 | c.598C>T/c.340+770T>C | VHL p.Arg200Trp/X1 p.Ser179Pro? | 1957/21 y | M | 19.9 | 67 | 22.2 | Erythrocytosis | — | |
| F3, II.1 | c.598C>T/c.340+770T>C | VHL p.Arg200Trp/X1 p.Ser179Pro? | 2003/4 mo | M | 17.9 | 54 | 7.5 | 60 | Erythrocytosis | — |
| F3, II.2 | c.598C>T/c.340+770T>C | VHL p.Arg200Trp/X1 p.Ser179Pro? | 2005/6 mo | M | 14.6 | 45 | Erythrocytosis | — | ||
| F4, II.1 | c.598C>T/c.340+694_711dup | VHL p.Arg200Trp/X1 p.Trp159X? | 1990/7 y | M | 20.6 | 64 | 8.25 | 49.9 | Erythrocytosis, deep vein thrombosis, intracerebral hemorrhage | FV Leiden |
| F5, II.2 | c.430G>A/c.340+694_711dup | VHL p.Gly144Arg/X1 p.Trp159X? | 1975/13 y | M | 16.2 | 60 | 5.9 | Erythrocytosis | — | |
| F6, II.1 | c.492G>T/c.340+574A>T* | VHL p.Gln164His/SA | 2014/6 mo | M | 15.6 | 49 | 6.9 | 1167 | Splenomegaly | — |
| F7 | c.340+816A>C/c.340+816A>C | X1 p.*194Serext*24/X1 p.*194Serext*24 | 7 y | M | 20 | 64 | 8.09 | 33 | Erythrocytosis | — |
| F8 | c.340+617C>G + c.340+648T>C†/WT | X1 p.Leu128Val+ X1 p.Leu138Pro/WT | N | N | N | N | VHL disease | — | ||
| F9, II.1 | c.429C>T/c.429C>T | VHL p.Asp143Asp/VHL p.Asp143Asp | 1997/5 mo | M | 16.2 | 49.3 | 5.2 | 186 | Erythrocytosis | Hb sitia |
| F10, II.1 | c.429C>T/c.429C>T | VHL p.Asp143Asp/VHL p.Asp143Asp | 2002/4.5 y | F | 22.5 | 64-77.4 | 12.6 | 264 | Erythrocytosis, splenomegaly | β thalassemia |
| F11 | c.414A>G/WT | VHL p.Pro138Pro/WT | N | N | N | N | VHL disease | — | ||
| F12 | c.414A>G/WT | VHL p.Pro138Pro/WT | N | N | N | N | VHL disease | — |
The second column indicates the position of the nucleotide variants identified in the VHL gene regarding the current nomenclature (sequence encoded by the VHL E1-E2-E3). The third column indicates the impact either on the VHL protein (for variants located in E1, E2, or E3) or on the potential X1 protein, if encoded by E1-E1′ (for variants located in E1′). Normal values correspond to: Hb: M, 13 to 18 g/dL and F, 12 to 15 g/dL; Ht: M, 40% to 52% and F, 37% to 47%; red cells: M, 4.2 to 5.7 × 109/L and F, 4.2 to 5.2 × 109/L; EPO: 5 to 25 mU/mL.
EPO, erythropoietin; Hb, hemoglobin; Ht, hematocrit; N, normal value.
The nucleotide change was reported as rs982745672 with a global minor allele frequency that corresponds to T = 0.00007/2 (TOPMed).
The nucleotide change was reported as rs73024533 with a global minor allele frequency that corresponds to C = 0.0026/13 (1000 Genomes) and C = 0.0050/147 (TOPMed).
New E1′ cryptic exon is mutated in patients with erythrocytosis or VHL disease
Sanger sequencing of this new cryptic exon in the proband (F1-II.1) identified a variant that had not been reported in databases: c.340+770T>C (Figure 2B; Table 1; supplemental Figure 4). Sequencing of the germline DNA of the mother revealed the same variant, indicating that the propositus F1-II.1 was compound heterozygous. This result prompted us to sequence additional patients with erythrocytosis described as heterozygous for VHL mutations.
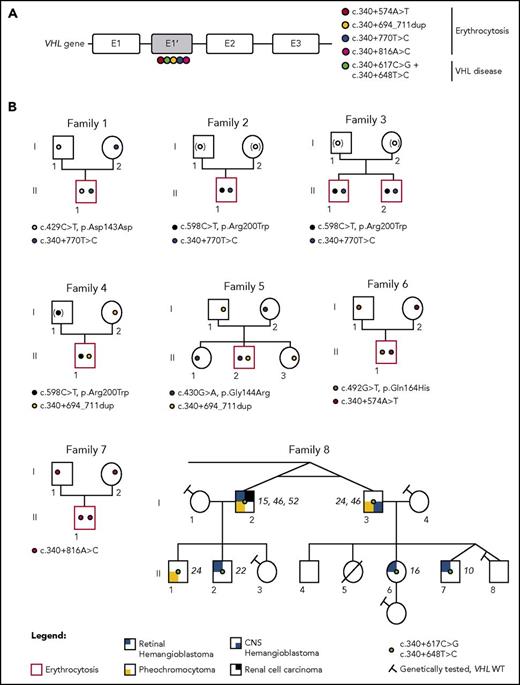
Identification of mutations in the new VHL cryptic exon in 7 families with erythrocytosis and a large family with VHL disease. (A) Schematic representation of the VHL gene and location of the identified mutations in the new VHL cryptic exon E1′. (B) Pedigree of families with erythrocytosis or VHL disease. The genotypes were elucidated by sequencing both parents and proband (F1, F5, and F6), deduced by sequencing of 1 parent and proband (F4; the mutation deduced in brackets), or deduced from allele cloning of proband carrying the conserved Chuvash mutation and core haplotype (F2 and F3), confirming the transmission of the mutations by 1 of each parent (for F2 and F3, the identity of the transmitting parent being unknown, the mutation is represented by a white circle in brackets). The genotype of parents from F7 was elucidated from WGS data. The numbers in italics (F8) indicate the age of the patient at tumor diagnosis. CNS, central nervous system.
Identification of mutations in the new VHL cryptic exon in 7 families with erythrocytosis and a large family with VHL disease. (A) Schematic representation of the VHL gene and location of the identified mutations in the new VHL cryptic exon E1′. (B) Pedigree of families with erythrocytosis or VHL disease. The genotypes were elucidated by sequencing both parents and proband (F1, F5, and F6), deduced by sequencing of 1 parent and proband (F4; the mutation deduced in brackets), or deduced from allele cloning of proband carrying the conserved Chuvash mutation and core haplotype (F2 and F3), confirming the transmission of the mutations by 1 of each parent (for F2 and F3, the identity of the transmitting parent being unknown, the mutation is represented by a white circle in brackets). The genotype of parents from F7 was elucidated from WGS data. The numbers in italics (F8) indicate the age of the patient at tumor diagnosis. CNS, central nervous system.
We investigated patients with Chuvash polycythemia for whom VHL R200W had been found in a heterozygous state. Sanger sequencing identified the identical intronic variant c.340+770T>C in a singleton (F2)5 and 2 affected brothers (F3; Figure 2B). In addition, we identified a duplication c.340+694_711dup in the proband of F4 and F56 previously described as VHL R200W/WT and VHL G144R/WT, respectively (Figure 2B). In F6, previously diagnosed as VHL Q164H/WT, we identified a genetic variant c.340+574A>T that altered the consensus SA site ag/TC of E1′. This variant is described as a rare polymorphism (rs98274567) in the National Center for Biotechnology Information database (Table 1).
Because biological material from the parents of F2 and F3 was not available, we cloned the intronic region containing the E1′ variant and the Chuvash core haplotype single-nucleotide polymorphism rs779808 associated with the VHL R200W mutation (described27 to be located downstream of E1′, in position c.340+1150). Segregation analysis (F4, F5, and F6) or Chuvash core haplotype analysis (F2 and F3) demonstrated that all patients were compound heterozygous, with 1 mutation being inherited from each parent (supplemental Figure 5).
In a parallel independent study, WGS was used to investigate patients with Chuvash polycythemia, heterozygous for the VHL R200W mutation (F2 and F3). The intronic variant c.340+770T>C was the only rare variant identified in the VHL gene in these patients. Other WGS filtering strategies (supplemental Methods) did not identify any additional significant mutations in biologically relevant genes (supplemental Figure 6). This study also conducted WGS for a trio with congenital erythrocytosis for which prior whole-exome sequencing had not identified any mutations (F7). No rare biologically relevant variants were identified using the filtering strategy described in supplemental Methods. However, further inspection of the new cryptic exon led to the identification of a c.340+816A>C variant in the homozygous state in the proband, with both parents being heterozygous for this mutation.
In addition to these patients with erythrocytosis, in which no variants in VHL were initially detected, screening of patients with VHL disease may also fail to detect mutations in canonical VHL exons. An example of such a family (F8) with hereditary hemangioblastoma, clear cell renal cell carcinoma, and pheochromocytoma was studied. Microsatellite analysis demonstrated a cosegregation of markers surrounding the VHL region with the disease (supplemental Figure 7). Sequencing of the new E1′ cryptic exon in this family identified 2 heterozygous variants in E1′: a previously unreported c.340+617C>G variant and a c.340+648T>C variant described as a rare polymorphism in databases (Table 1). These variants segregated in 6 patients who developed the disease and were absent in 4 healthy tested descendants, indicating their presence on a single disease-associated allele (Figure 2B). Sequencing of tumor DNA did not display loss of heterozygosity, suggesting that a WT VHL deletion (as observed in classic VHL disease) may not be a prerequisite in cells of patients with this specific VHL genotype (supplemental Figure 4).
Expression study of the new VHL isoforms in patients’ cells
RNA sequencing (RNA-seq) analysis of patients’ LCLs and tumors demonstrated that genomic variants in E1′ were associated with an upregulation of transcripts containing the E1′ cryptic exon compared with controls (Figure 3A; supplemental Figure 8). In addition, the mutated allele was preferentially expressed (>70%), suggesting a causal relationship between the genetic variants and E1′ retention (supplemental Figure 8C).
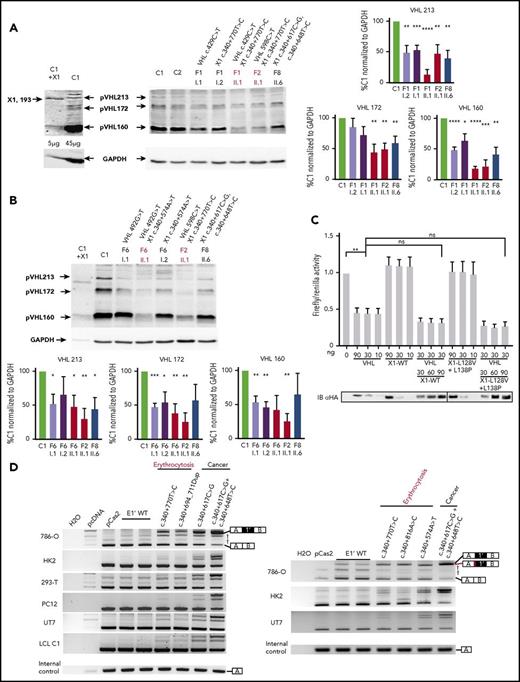
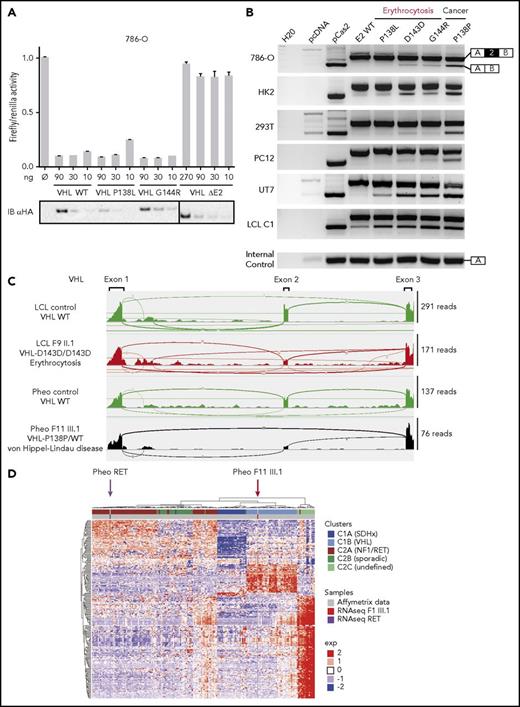
Expression study of the new VHL transcript isoforms in patient cells. (A) Sashimi plots from RNA-seq data obtained from samples (LCLs or pheochromocytomas [Pheo]) of 3 patients from F8. The positions of the different VHL exons are indicated, with the maximal number of reads indicated at the right. Splice junctions are denoted by the horizontal links, with details provided in supplemental Figure 8. (B) TaqMan quantification of the different VHL isoforms from samples of F8 performed using probes specific to VHL E1-E2, the translated sequence upstream E1′, or E1-E1′ junctions. Relative gene expression was normalized to LCL control (C1) fixed at 1 (mean results of technical duplicates). (C) TaqMan quantification of the different VHL isoforms in LCLs (established from 2 independent controls and from patients of F1, F2, and F8) cultured in the absence or presence of puromycin (puro), an inhibitor of nonsense-mediated mRNA decay. TaqMan probes were specific to the VHL E1-E1′ or E1-E2 junction. The graph resumes experiments performed on LCLs cultured independently 3 times and quantified in duplicate. Data are represented as the mean ± standard error of the mean. (D) RT-PCR using primers specific for E1 and E3 was performed on mRNA extracted from LCLs established from controls and patients of F6 (carrying the mutation c.340+574A>T that targets the SA site of E1′). Patient with erythrocytosis is indicated in red. On the right, the spliced isoforms are schematically represented. **P < .05, ***P < .01 based on Student t test. *Denotes larger fragments that contained E1′ spliced with E1, but with 15 nucleotides deleted (represented in red) by the use of an alternative SA site (sequences of the cloned bands are detailed in supplemental Figure 10).
Expression study of the new VHL transcript isoforms in patient cells. (A) Sashimi plots from RNA-seq data obtained from samples (LCLs or pheochromocytomas [Pheo]) of 3 patients from F8. The positions of the different VHL exons are indicated, with the maximal number of reads indicated at the right. Splice junctions are denoted by the horizontal links, with details provided in supplemental Figure 8. (B) TaqMan quantification of the different VHL isoforms from samples of F8 performed using probes specific to VHL E1-E2, the translated sequence upstream E1′, or E1-E1′ junctions. Relative gene expression was normalized to LCL control (C1) fixed at 1 (mean results of technical duplicates). (C) TaqMan quantification of the different VHL isoforms in LCLs (established from 2 independent controls and from patients of F1, F2, and F8) cultured in the absence or presence of puromycin (puro), an inhibitor of nonsense-mediated mRNA decay. TaqMan probes were specific to the VHL E1-E1′ or E1-E2 junction. The graph resumes experiments performed on LCLs cultured independently 3 times and quantified in duplicate. Data are represented as the mean ± standard error of the mean. (D) RT-PCR using primers specific for E1 and E3 was performed on mRNA extracted from LCLs established from controls and patients of F6 (carrying the mutation c.340+574A>T that targets the SA site of E1′). Patient with erythrocytosis is indicated in red. On the right, the spliced isoforms are schematically represented. **P < .05, ***P < .01 based on Student t test. *Denotes larger fragments that contained E1′ spliced with E1, but with 15 nucleotides deleted (represented in red) by the use of an alternative SA site (sequences of the cloned bands are detailed in supplemental Figure 10).
We performed quantitative RT-PCR using TaqMan probes specific for the different splicing junctions or for the region upstream of E1′ (location of probes shown in Figure 1C), which showed a strong upregulation of a unique isoform containing E1′ spliced with E1 in LCLs and tumors from patients of F8 compared with controls (Figure 3B). The study therefore focused on the isoform E1-E1′. Transcripts that retained E1′ spliced with E1 contained (in-frame with E1) a premature termination codon (Figure 1C) and could likely be targeted for degradation according to nonsense-mediated mRNA decay (NMD) mechanisms. We therefore investigated the expression of transcripts that contained E1 spliced with E1′ in the absence or presence of puromycin, an inhibitor of NMD. Without puromycin, higher levels of expression were seen in samples with mutated E1′ (Figure 3C) compared with samples containing WT E1′. Exposure to puromycin resulted in a profound induction of isoforms containing E1 spliced with E1′ vs other isoforms (Figure 3C; supplemental Figure 9). These results indicated that isoforms with E1-E1′ junction were degraded by NMD and may have failed to produce proteins. We performed RT-PCR on LCL samples of F6 carrying the mutation in the SA site, using primers in VHL exons E1 and E3. We observed fragments of larger size in samples from patients carrying the mutation, which were highly visible in the presence of puromycin (Figure 3D). Cloning and sequencing of these fragments demonstrated a dysregulated splicing of E1′ with the use of an alternative SA site located 15 nucleotides downstream (supplemental Figure 10).
To measure the impact of the splicing dysregulation on pVHL expression, we performed an immunoblot with an antibody able to recognize the amino acids encoded by E1 and therefore able to detect the different pVHL isoforms.8 The antibody detected overexpressed exogenous X1 protein after transfection of the plasmid encoding X1 (Figure 4A). However, it failed to detect the endogenous protein, even in patients’ samples that overexpressed the mRNA containing E1-E1′. Instead, the immunoblots showed a lower expression of all the VHL protein isoforms in patients with mutated E1′ (Figures 4A-B).
Functional studies of mutations in VHL E1′. (A-B) Immunoblot analysis of patient LCLs. A representative immunoblot (A) and quantification of 3 different immunoblots (B) are presented. Relative gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and results obtained with LCL control (C1) were fixed at 100%. Data are represented as the mean ± standard error of the mean. A VHL antibody that recognized VHL E1 was able to detect the hypothetical X1 protein. In the most left lane, control LCLs were transfected with an expression vector containing the coding sequence for X1; 5 µg of proteins were blotted vs 45 µg for other samples to avoid signal saturation with overexpressed X1. Patients with erythrocytosis are indicated in red. (C) Functional hypoxia response element (HRE)–dependent reporter assays were performed in 786-O cells (ie, VHL− cells that constitutively express HIF-2α). The results are expressed as relative firefly luciferase activity with Renilla luciferase as an internal control; 1.0 unit denotes the basal activity of endogenous HIF-2α using the HRE luciferase reporter plasmid. The ability of WT and mutated X1 to downregulate firefly luciferase activity (related to HIF activity) was compared with pVHL and in competition with pVHL. An immunoblot using an antibody specific to the hemagglutinin tag (HA) was used to detect HA-VHL and HA-X1. X1-L128V+L138P corresponds to a potential impact of the c.340+648T>C and c.340+617C>G mutations on the hypothetical X1 protein. Three independent experiments were performed. (D) Characterization of VHL E1′ retention by the minigene experiment (representative picture of agarose gel; n = 3). RT-PCR was performed on mRNA obtained from cell lines transfected with a minigene construct containing VHL E1′ (WT or mutated) flanked by large intronic sequences cloned between the SERPING1 exons (exons A and B, targeted by the RT-PCR primers). The plasmids were transfected, and the expression of the spliced chimeric transcripts (containing EA and EB ± E1′) was analyzed. Two WT constructs containing E1′ were used; 1 contained the single-nucleotide polymorphism c.340+1150T>C (rs779808). Bands corresponding to EA and EB spliced together or with VHL E1′ are indicated on the right. *P < .05, **P < .01, ***P < .001, ****P < .0001 based on Student t test. †Corresponds to unspecific bands verified by sequencing. The minigene experiment performed with the construction carrying the mutation c.340+574A>T (that targets the SA site of E1′ in F6) confirmed the use of an alternative SA site (right panel) with the deletion of 15 nucleotides (represented in red). ns, not significant.
Functional studies of mutations in VHL E1′. (A-B) Immunoblot analysis of patient LCLs. A representative immunoblot (A) and quantification of 3 different immunoblots (B) are presented. Relative gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and results obtained with LCL control (C1) were fixed at 100%. Data are represented as the mean ± standard error of the mean. A VHL antibody that recognized VHL E1 was able to detect the hypothetical X1 protein. In the most left lane, control LCLs were transfected with an expression vector containing the coding sequence for X1; 5 µg of proteins were blotted vs 45 µg for other samples to avoid signal saturation with overexpressed X1. Patients with erythrocytosis are indicated in red. (C) Functional hypoxia response element (HRE)–dependent reporter assays were performed in 786-O cells (ie, VHL− cells that constitutively express HIF-2α). The results are expressed as relative firefly luciferase activity with Renilla luciferase as an internal control; 1.0 unit denotes the basal activity of endogenous HIF-2α using the HRE luciferase reporter plasmid. The ability of WT and mutated X1 to downregulate firefly luciferase activity (related to HIF activity) was compared with pVHL and in competition with pVHL. An immunoblot using an antibody specific to the hemagglutinin tag (HA) was used to detect HA-VHL and HA-X1. X1-L128V+L138P corresponds to a potential impact of the c.340+648T>C and c.340+617C>G mutations on the hypothetical X1 protein. Three independent experiments were performed. (D) Characterization of VHL E1′ retention by the minigene experiment (representative picture of agarose gel; n = 3). RT-PCR was performed on mRNA obtained from cell lines transfected with a minigene construct containing VHL E1′ (WT or mutated) flanked by large intronic sequences cloned between the SERPING1 exons (exons A and B, targeted by the RT-PCR primers). The plasmids were transfected, and the expression of the spliced chimeric transcripts (containing EA and EB ± E1′) was analyzed. Two WT constructs containing E1′ were used; 1 contained the single-nucleotide polymorphism c.340+1150T>C (rs779808). Bands corresponding to EA and EB spliced together or with VHL E1′ are indicated on the right. *P < .05, **P < .01, ***P < .001, ****P < .0001 based on Student t test. †Corresponds to unspecific bands verified by sequencing. The minigene experiment performed with the construction carrying the mutation c.340+574A>T (that targets the SA site of E1′ in F6) confirmed the use of an alternative SA site (right panel) with the deletion of 15 nucleotides (represented in red). ns, not significant.
Functional characterization of the mutated VHL E1′
Because the hypoxia pathway represents the major pathway involved in the genesis of secondary erythrocytosis and VHL disease,13-15,28 we performed functional studies of the hypothetical WT or mutated X1 proteins using hypoxia response element–dependent reporter assays. These functional tests failed to reveal any substantial effects of X1 on this pathway, either alone or in competition with pVHL (Figure 4C; supplemental Figure 11). We next focused our study on a potential impact of the E1′ variants on splicing by performing splicing reporter minigene assays in various cell lines. The experiments showed that splicing of WT E1′ was barely detected (Figure 4D). Interestingly, higher molecular weight bands corresponding to the expected spliced isoforms containing E1′ appeared in the presence of the mutations. Cloning and sequencing of the isoforms confirmed a retention of E1′ at the expected splicing sites. The level of expression of this upper isoform, specific to E1′ inclusion, was higher for mutations associated with cancers than for those with erythrocytosis, independently of the cell lines. Notably, the combination of both variants associated with cancer had a more pronounced effect than each variant individually (the single-nucleotide polymorphism showing very low effect), suggesting a synergistic effect of the variants on splicing (supplemental Figure 12). In silico analysis of c.340+617C>G consistently predicted a severe alteration of splicing by the creation of an exonic splicing enhancer site (supplemental Figure 13).
The minigene experiment performed with mutated E1′ at the SA site (F6) confirmed the inclusion of E1′ during splicing (Figure 4D, right panel), using the same downstream alternative SA site identified in mRNA extracted from patients’ LCLs (supplemental Figure 10).
Synonymous mutations in VHL E2 induce exon skipping
We then focused our study on the synonymous heterozygous D143D mutation in E2, identified in the proband of F1. This mutation was also identified in the homozygous state in 2 patients with erythrocytosis (F9 and F10; Figure 5A; Table 1). The mRNA extracted from LCLs established from the different members of these 2 families was reverse transcribed and sequenced. Comparison of chromatograms obtained by sequencing of DNA vs cDNA displayed a weaker peak of the mutated allele in cDNA, reflecting a decreased expression of the mRNA transcripts carrying the mutation.
Genetic and expression study of synonymous mutations in VHL E2. (A-B) Pedigree and sequence chromatograms of germline DNA, tumor (pheochromocytoma) DNA, or cDNA prepared from 2 families (F9 and F10) with erythrocytosis (A) and 2 families (F11 and F12) with VHL disease (B). (C) Results of RT-PCR using mRNA extracted from LCLs (F9 and F10) and leukocytes and tumor material (pheochromocytoma) (F11 and F12). (D) TaqMan quantification of the different VHL isoforms in LCLs (established from patients of F9, F10, and F11) cultured in the absence or presence of puromycin. TaqMan probes are specific to the VHL E1-E2 or E1-E3 junction. Relative gene expression was normalized to LCL control (C1). (E) Immunoblot analysis of patient LCLs. Patients with erythrocytosis are indicated in red. A representative immunoblot (upper panel) and quantification of 4 different immunoblots are displayed (lower panel). Relative gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and results obtained with C1 were fixed at 100%. Data are represented as the mean ± standard error of the mean. *P < .05, **P < .01, ***P < .001, ****P < .0001 based on Student t test. CNS, central nervous system.
Genetic and expression study of synonymous mutations in VHL E2. (A-B) Pedigree and sequence chromatograms of germline DNA, tumor (pheochromocytoma) DNA, or cDNA prepared from 2 families (F9 and F10) with erythrocytosis (A) and 2 families (F11 and F12) with VHL disease (B). (C) Results of RT-PCR using mRNA extracted from LCLs (F9 and F10) and leukocytes and tumor material (pheochromocytoma) (F11 and F12). (D) TaqMan quantification of the different VHL isoforms in LCLs (established from patients of F9, F10, and F11) cultured in the absence or presence of puromycin. TaqMan probes are specific to the VHL E1-E2 or E1-E3 junction. Relative gene expression was normalized to LCL control (C1). (E) Immunoblot analysis of patient LCLs. Patients with erythrocytosis are indicated in red. A representative immunoblot (upper panel) and quantification of 4 different immunoblots are displayed (lower panel). Relative gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and results obtained with C1 were fixed at 100%. Data are represented as the mean ± standard error of the mean. *P < .05, **P < .01, ***P < .001, ****P < .0001 based on Student t test. CNS, central nervous system.
A different synonymous mutation in E2, c.414A>G p.Pro138Pro (P138P), was identified in 2 families (F11 and F12) with VHL disease (Figure 5B). This heterozygous mutation segregated with the disease in 3 generations. Sequencing of DNA extracted from the pheochromocytoma of F11-III.1 showed a loss of the WT allele in the tumor, demonstrating loss of heterozygosity as currently described in classical VHL disease.
Suspecting an effect of the synonymous mutations on splicing, we next assessed the expression of the VHL transcripts in patients’ samples by RT-PCR (Figure 5C). We found a significant change in the ratio of expressed VHL isoforms, with a higher expression of the E1-E3 transcripts in LCLs of patients homozygous for D143D and in the pheochromocytoma with P138P mutation. These results suggest an effect of the mutation on splicing regulatory elements, leading to E2 skipping. In silico analysis of the synonymous mutations indicated a potential effect on exonic splicing enhancer motifs (supplemental Figure 14).
To specifically quantify the different VHL isoforms, we performed quantitative RT-PCR on LCLs using TaqMan probes complementary to the VHL E1-E2 or E1-E3 junction (Figure 5D). All mutated samples displayed an increase of the isoform with skipped E2 (E1-E3). The patients carrying a homozygous D143D mutation presented a severe decrease of the WT isoform expression level. These results demonstrated that synonymous mutations in E2 affected mRNA isoform production.
To evaluate a potential impact on protein expression, we performed a western blot analysis. We did not observe any overexpression of the pVHL172 isoform as expected from mRNA quantification studies. However, in patients homozygous for D143D, we detected a strong downregulation of all the pVHL isoforms, which was also observed to a lesser extent in LCL samples from heterozygous patients (Figure 5E).
Proportion of VHL E2 skipping is correlated with disease severity in minigene experiments
To study the impact of the synonymous E2 mutations on splicing in different cell lines, we performed minigene assays. We tested the implications of the D143D and P138P mutations on splicing in addition to the nearby mutations described in patients with erythrocytosis (P138L and G144R).4,6,29 Indeed, these mutations may also affect splicing rather than VHL protein function. We first evaluated the loss of function of these mutants by reporter assays. We found that these could downregulate HIF in a manner similar to the WT protein, in contrast to VHL proteins lacking E2 (pVHLΔE2), which are described as nonfunctional in terms of their regulation of HIF activity8-10,12 (Figure 6A). Minigene experiments were performed in a variety of cell lines relevant to the studied diseases, using LCLs as control (Figure 6B). We demonstrated that all mutations caused E2 skipping in LCLs. In other cell lines, these experiments demonstrated a major impact on the VHL E2 splicing of the P138P mutation, which is associated with cancer. The mutations P138L, D143D, and G144R, which are all associated with erythrocytosis, displayed a weaker effect on splicing, with slight variations among cell lines but with a stronger effect in the erythroid cell line (UT7 cultured with erythropoietin).
Functional study of synonymous mutations in VHL E2. (A) Functional hypoxia response element (HRE)–dependent reporter assays were performed in 786-O cells to evaluate the impact of VHL mutations in E2 (P138L and G144R) on VHL protein activity. The VHL protein lacking E2 (pVHL172/VHLΔE2) was used as a negative control. The results are expressed as firefly luciferase activity relative to Renilla luciferase as an internal control; 1.0 unit denotes the basal activity of endogenous HIF-2α using the HRE luciferase reporter plasmid. Immunoblots using an antibody specific for the hemagglutinin tag (HA) were used to detect HA-VHL. (B) Characterization of VHL E2 skipping by minigene analyses. Minigene experiments were performed in a variety of cell lines relevant to the studied diseases: renal (293T, 786-O, and HK2), pheochromocytoma (pheo; PC12), erythroid cell line (UT7 cultured with erythropoietin), and LCL. RT-PCR was performed on mRNA obtained from cell lines transfected with a minigene construct containing VHL E2 flanked by intronic sequences cloned between the SERPING1 exons (exons A and B; targeted by the RT-PCR primers). Bands corresponding to EA and EB spliced together or with VHL E2 are indicated on the right. (C) Sashimi plots from RNA-seq data. The positions of the different VHL exons are indicated, with the maximum number of reads for each exon indicated at the right. Splice junctions are denoted by the horizontal links, with details provided in supplemental Figure 15A. (D) Heatmap of pheochromocytoma transcriptome data. A comparison of transcriptome data for the pheochromocytoma from patient F11 III.1 (with P138P mutation) vs Affymetrix data from the largest available cohort of paragangliomas/pheochromocytomas (recruited by the French COMETE network) that identified homogeneous molecular subgroups associated with susceptibility genes (Burnichon et al30 ).
Functional study of synonymous mutations in VHL E2. (A) Functional hypoxia response element (HRE)–dependent reporter assays were performed in 786-O cells to evaluate the impact of VHL mutations in E2 (P138L and G144R) on VHL protein activity. The VHL protein lacking E2 (pVHL172/VHLΔE2) was used as a negative control. The results are expressed as firefly luciferase activity relative to Renilla luciferase as an internal control; 1.0 unit denotes the basal activity of endogenous HIF-2α using the HRE luciferase reporter plasmid. Immunoblots using an antibody specific for the hemagglutinin tag (HA) were used to detect HA-VHL. (B) Characterization of VHL E2 skipping by minigene analyses. Minigene experiments were performed in a variety of cell lines relevant to the studied diseases: renal (293T, 786-O, and HK2), pheochromocytoma (pheo; PC12), erythroid cell line (UT7 cultured with erythropoietin), and LCL. RT-PCR was performed on mRNA obtained from cell lines transfected with a minigene construct containing VHL E2 flanked by intronic sequences cloned between the SERPING1 exons (exons A and B; targeted by the RT-PCR primers). Bands corresponding to EA and EB spliced together or with VHL E2 are indicated on the right. (C) Sashimi plots from RNA-seq data. The positions of the different VHL exons are indicated, with the maximum number of reads for each exon indicated at the right. Splice junctions are denoted by the horizontal links, with details provided in supplemental Figure 15A. (D) Heatmap of pheochromocytoma transcriptome data. A comparison of transcriptome data for the pheochromocytoma from patient F11 III.1 (with P138P mutation) vs Affymetrix data from the largest available cohort of paragangliomas/pheochromocytomas (recruited by the French COMETE network) that identified homogeneous molecular subgroups associated with susceptibility genes (Burnichon et al30 ).
Splicing dysregulation of VHL is causal in the development of disease
RNA-seq confirmed that the synonymous mutations were not silent but instead induced potent E2 skipping (Figure 6C; supplemental Figure 15A). Additional transcriptome analyses of the pheochromocytoma carrying P138P showed an upregulation of HIF target genes (typically seen in VHL-related pheochromocytomas) compared with the pheochromocytoma carrying a RET mutation (supplemental Figure 15B).30 We then reanalyzed our independent cohort of pheochromocytomas30 using RNA-seq data from both tumors. After unsupervised classification, we observed a segregation of the P138P pheochromocytoma with other VHL-related tumors (C1B cluster), whereas the control pheochromocytoma bearing the RET mutation was grouped with other RET-related tumors (Figure 6D).
Discussion
The hypoxia pathway plays a central role in erythrocytosis or tumors developed by patients carrying VHL mutations. Nevertheless, the full molecular mechanisms at the origin of these different phenotypes remain to be elucidated. To date, functional studies of VHL mutants have been performed on missense mutations. We describe here, for the first time, functional studies of VHL mutations that did not have an impact on the coding sequence but that influenced VHL splicing. We discovered a complex regulation of VHL splicing that may help to explain the complexity of genotype/phenotype correlations observed in VHL-related disorders. Notably, we demonstrated that synonymous variants (D143D or P138P) could have an impact on VHL splicing and should be considered as pathogenic mutations. Our study points to a particular region in E2 that may be considered as a splicing regulatory domain. Therefore, it would be interesting to evaluate the impact of all the nucleotide changes described in VHL E217 on splicing, in the same way as described for 2 missense mutations (P138L and G144R). We observed that, depending on the mutation in this region, the impact on splicing could be moderate (D143D, G144R, and P138L) or severe (P138P), correlating with the severity of the disease seen in individuals carrying these VHL mutations (erythrocytosis vs cancers). This observation confirms the hypothesis of a continuum model of tumor suppression by VHL.21,31
Regarding the erythrocytosis developed in patients homozygous for D143D, both probands (F9 II.1 and F10 II.1) presented mutations in the β-globin gene (HBB) that induced hemoglobin instability or thalassemia (Table 1). This may have compensated for the strong erythropoiesis associated with a very high serum erythropoietin level associated with the D143D mutation.
More importantly, we discovered a new VHL cryptic exon, E1′, expressed in healthy tissues. Our study led to the identification of E1′ heterozygous mutations occurring in the second allele of 6 families with erythrocytosis previously associated with a heterozygous mutation in VHL rather than a homozygous mutation. Our investigations further confirmed that polycythemia associated with VHL mutation was definitely an autosomal recessive disease. In addition, we identified an E1′ homozygous mutation in a patient with erythrocytosis of unknown origin. This result demonstrated the causal role of the alterations in this new cryptic exon in the pathophysiology of erythrocytosis. Importantly, we also identified E1′ mutations in patients with unexplained VHL disease.
This VHL E1′ exon remained unidentified until this point because of its low expression and the fact that this deep intronic region was never explored or represented in whole-exome sequencing data. Our data showed that these newly described E1′-containing transcripts may have been polyadenylated (because they were captured by polydT in RNA-seq) but were also likely to have been subjected to NMD and may therefore have failed to produce a protein. However, we cannot exclude the possibility that they were translated into a new protein (X1) not expressed in sufficient quantity to be detected by western blot. This potential X1 isoform would contain the VHL E1 that encodes the NH2-terminal part of pVHL, including 16 residues involved in HIF binding from the 17 described.26 Nonetheless, the hypoxia-dependent reporter assays failed to identify a potential direct role of this isoform in the HIF pathway. Instead, our results provided compelling evidence that mutations in E1′ induced a severe retention of this E1′ cryptic exon, which correlated with a defect in global VHL protein expression. Therefore, our study strongly favors a dysregulation of splicing with a consequent downregulation of pVHL expression as the underlying cause of the diseases observed. Here, insufficient pVHL levels but not reduced HIF binding by the mutant pVHL (as seen in the Chuvash polycythemia mutation VHL R200W1 ) may have led to an impairment of HIF degradation. Of note is the fact that the functional study of mutations identified in E1′ in association with erythrocytosis resulted in a less severe impact on splicing than the mutations associated with cancer, confirming that polycythemia was associated with VHL hypomorphic mutations.
Our findings may have broad implications for patients with presumed congenital erythrocytosis. First, the underlying cause of congenital erythrocytosis has been identified in only approximately one third of patients in most published studies. However, previous studies have focused on missense and nonsense changes in coding regions and known regulatory domains of candidate genes. This has also been the case in patients with suspected VHL disease. Our study shows that synonymous exonic changes, as well as changes within intronic sequences affecting exon splicing, may be responsible for these disorders and should be considered during the diagnostic process. Notably, the VHL E1′ exon should be added to the list of regions routinely sequenced in patients with congenital erythrocytosis. Second, the detection of molecular changes has implications for the clinical management of patients. For example, phlebotomy in patients with VHL- or HIF2A–related erythrocytosis may worsen the clinical situation by increasing the risk and severity of pulmonary hypertension in these patients.32 Third, the confirmation of the continuum model of tumor suppression by VHL helps us to understand the very low frequency of secondary tumors in patients with VHL-related erythrocytosis.33 However, this also means that later occurrence of such neoplasms cannot be definitely excluded. Therefore, the clinical management of these patients should include regular follow-up to assess the risk for thromboembolic complications, pulmonary hypertension, and cardiovascular disease, as well as regular examinations to check for the presence of typical VHL-related tumors. Patients with VHL disease resulting from E1′ mutations will also benefit from regular screening for tumors. Finally, the detection of these new genetic changes will also allow for appropriate genetic counseling of affected patients and their families.
In conclusion, VHL is a major tumor suppressor gene that plays a pivotal role in the oxygen-sensing pathway, which is involved in multiple physiological (eg, angiogenesis and erythropoiesis) and pathological (eg, cancer) processes. Our findings regarding the complex splicing regulation of this gene in erythrocytosis and tumorigenesis may therefore open new avenues for diagnosis of these conditions as well as research on the biological mechanisms related to the hypoxia-signaling pathway. Notably, we suggest further targeted exploration of the VHL E1′ region in unresolved cases of congenital erythrocytosis, inherited kidney cancers, hereditary paraganglioma/pheochromocytoma syndrome, and hemangioblastomas, in addition to all types of sporadic tumors with altered hypoxia signaling.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Richard Breathnach, Jean Feunteun, Judith Favier, and Sylvie Hermouet for scientific discussions; Helena M. Silva, Caroline Abadie, Sophie Giraud, Florence Fellmann, Pascal Pigny, Vinciane Dideberg, and Segers Karine for patient recruitment and medical advice; and Amandine Le Roy, Isabelle Barbieux, Christophe Simian, Marie-Laure Clenet, and Stéphanie Lebeau for technical assistance, in addition to the Genomics and Bioinformatics Core Facility of Nantes (GenoBiRD; Biogenouest) for its technical support. The authors are grateful to Mario Tosi, Pascaline Gaildrat, and Alexandra Martins (Institute for Research and Innovation in Biomedicine, INSERM U1079, University of Rouen, Rouen, France) for kindly providing the pCAS2 minigene plasmid. The authors thank the European Cooperation in Science and Technology (COST) Networks Molecular Diagnosis of Myeloproliferative Neoplasms (MPNs) and MPN-Related Congenital Diseases Euronet (COST action BM0902) and Hypoxia Sensing, Signaling, and Adaptation Hypoxianet (COST action TD0901). The authors also thank the Patients' Association VHL France for its constant support.
This study was supported by grants from the Région Pays de la Loire, Project EryCan; the Agence Nationale Recherche (Programme de Recherche Translationnelle en Santé 2015 GenRED); the Laboratory of Excellence GR-Ex (reference #ANR-11-LABX-0051); and the Ligue Nationale Contre le Cancer (Comités de la Loire Atlantique et des Côtes d’Armor). In addition, we acknowledge funding from the Oxford National Institute for Health Research Biomedical Research Centre and the Health Innovation Challenge Fund scheme. The views expressed in this manuscript are those of the authors and not necessarily those of the Wellcome Trust and Department of Health. We also acknowledge funding from the Wellcome Trust Core Award (grant #203141/Z/16/Z). A.B. received financial support from L’Institut Thématique Multi-Organisme Cancer Alliance Nationale pour les Sciences de la Vie et de la Santé (National Alliance for Life Sciences and Health) within the framework of the Cancer Plan. D.H. is supported by the National Center of Competence in Research Kidney Control of Homeostasis.
Authorship
Contribution: M.L., F.R., K.S., A.C., D.H., A.B., S.G., S.C., F.C., M.P., T.B., S.F., M.V., Y.A.-B., and B.G. performed experiments; C.C., S.J.L.K., J.C.T., M.P., E.K., and H.D. conducted genome sequencing or data analysis and interpretation; P.L., M.C., S. Dumont, S.J., and M.L. performed bioinformatic analyses; H.C., K.S., S. Deveaux, N.B., J.-M.M., F.A., C.G., L.H., S.I., E.M., K.B., K.-M.D., B.B.-d.P., F.G., M.-L.R., M.C.P., V.B., R.V.W., J.R.G., A.K., N.J., C.B., A.-P.G.-R., and S.R. conducted the medical study; B.G., D.H., M.L., and F.R. wrote the manuscript; B.G. and H.C. designed the study; B.G. directed the study; and all authors contributed to the research and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Betty Gardie, Ecole Pratique des Hautes Etudes, L'Institut du Thorax, INSERM, CNRS, Université de Nantes, Nantes, France; e-mail: betty.gardie@inserm.fr.
REFERENCES
Author notes
M.L. and F.R. contributed equally to this work.
S.R. and A.-P.G.-R. contributed equally to this work.
H.C. and B.G. contributed equally to this work.

![Figure 3. Expression study of the new VHL transcript isoforms in patient cells. (A) Sashimi plots from RNA-seq data obtained from samples (LCLs or pheochromocytomas [Pheo]) of 3 patients from F8. The positions of the different VHL exons are indicated, with the maximal number of reads indicated at the right. Splice junctions are denoted by the horizontal links, with details provided in supplemental Figure 8. (B) TaqMan quantification of the different VHL isoforms from samples of F8 performed using probes specific to VHL E1-E2, the translated sequence upstream E1′, or E1-E1′ junctions. Relative gene expression was normalized to LCL control (C1) fixed at 1 (mean results of technical duplicates). (C) TaqMan quantification of the different VHL isoforms in LCLs (established from 2 independent controls and from patients of F1, F2, and F8) cultured in the absence or presence of puromycin (puro), an inhibitor of nonsense-mediated mRNA decay. TaqMan probes were specific to the VHL E1-E1′ or E1-E2 junction. The graph resumes experiments performed on LCLs cultured independently 3 times and quantified in duplicate. Data are represented as the mean ± standard error of the mean. (D) RT-PCR using primers specific for E1 and E3 was performed on mRNA extracted from LCLs established from controls and patients of F6 (carrying the mutation c.340+574A>T that targets the SA site of E1′). Patient with erythrocytosis is indicated in red. On the right, the spliced isoforms are schematically represented. **P < .05, ***P < .01 based on Student t test. *Denotes larger fragments that contained E1′ spliced with E1, but with 15 nucleotides deleted (represented in red) by the use of an alternative SA site (sequences of the cloned bands are detailed in supplemental Figure 10).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/5/10.1182_blood-2018-03-838235/4/m_blood838235f3.jpeg?Expires=1769122423&Signature=rJKiz-3QCR87Z7K1CZldCOrizyMKWKWFGhEnUd2wkeQMeAjYyTxe~LlZimn8J7zJ1p2oiYHsVD1hSkqfIeOFq9Ns2Y2h9mL98LV-jA0ucBXgkcGwzSR5fbJsTr-oHiTP3kaDzAXdzMWfb6J9uwIamLDakpE-PZRiPeHCAijkFmvaVEKtJaWjKLRtGVG9NedCW3kkHlHzh973raFSQKI2w-iuCjFXHnXCMJZtWQTbJDxV3apKbVKSIURvAErgIoIQhktwUUYzaQIs6zkDBj9~sJ4dBo2okMHJ5s7pZ1gHvs93ENQDL78vojrgA7H25LMRyQS2CQ81jdoeavCZryWk1g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal