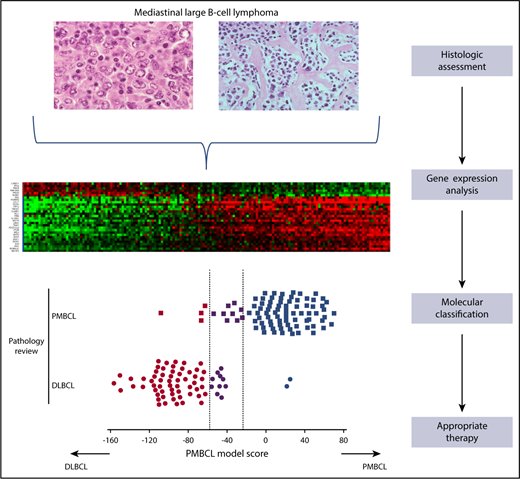
Use of a gene expression-based assay in the molecular classification of large B-cell lymphoma in the mediastinum into PMBCL and DLBCL.
Use of a gene expression-based assay in the molecular classification of large B-cell lymphoma in the mediastinum into PMBCL and DLBCL.
PMBCL is a unique clinicopathologic subtype of aggressive B-cell lymphoma, the diagnosis of which is dependent on correlating clinical features and pathologic and immunophenotypic features.2 Despite improvements in the diagnostic criteria that have been developed for many subtypes of non-Hodgkin lymphoma, distinguishing PMBCL from DLBCL and gray zone lymphoma involving the mediastinum is still a challenge when using currently available diagnostic methods.
By leveraging the wealth of gene expression profiling (GEP) data obtained from PMBCL samples,3,4 the authors sought to establish a molecular assay that would be broadly applicable for routine clinical diagnostic testing. Important criteria were the ability to use nucleic acids obtained from routinely fixed FFPETs and to use a widely available diagnostic platform. The authors addressed this challenge by developing a quantitative gene expression assay that analyzed 58 genes using the Nanostring platform labeled Lymph3Cx.
The Mottok article describes a rigorous study that used a training set composed of 68 cases of PMBCL and DLBCL and an independent validation set composed of 158 cases. The authors used 30 genes to distinguish between PMBCL and DLBCL, 24 of which were overexpressed in PMBCL and 6 of which were overexpressed in DLBCL and represented high discriminative power. The assay also included genes that are represented in Lymph2Cx, which allowed for cell-of-origin (COO) determination in DLBCL not otherwise specified (NOS). Thus, the assay could use molecular signatures to distinguish PMBCL and also provide COO assignments to DLBCL NOS (see figure). The reproducibility of the assay performance in 2 independent laboratories was high, which supports the likelihood of this assay becoming clinically relevant.
Patients with PMBCL demonstrate selective responses to novel therapeutic approaches such as dose-adjusted etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab (EPOCH-R)5 or anti-programmed cell death protein 1 antibody6 vs those therapies for DLBCL. Thus, the ability to more accurately distinguish PMBCL from DLBCL has important implications for patient management.
Potential obstacles in the rapid and large-scale adoption of this assay in routine clinical settings are the availability of the technical platform and the amount of tissue required (core needle biopsies vs excisional lymph node biopsies) for the assay to perform at the levels reported in the Mottok article. Most importantly, the ability to translate the results of the Mottok study is limited by the fact that the assay was developed by using a specific technical platform, and thus the applicability to other GEP methods such as reverse transcriptase multiplex ligation-dependent probe amplification7 is unknown.
One major diagnostic challenge that is not addressed is the distinction between PMBCL and classical Hodgkin lymphoma (CHL). Given the biologic overlap between these 2 entities, GEP assays such as those described in the Mottok article may provide a more robust and accurate molecular assay to distinguish PMBCL from CHL and provide quantitative gene expression-based cutoff values that could be used to assign diseases that truly represent gray zone lymphomas.
The Mottok study highlights the ongoing development of quantitative robust GEP-based molecular assays such as DLBCL COO classification8 and assays that may be in current development for classifying peripheral T-cell lymphoma.9 These add to the expanding armamentarium of molecular assays (eg, circulating tumor DNA, targeted next-generation sequencing panels, epigenetic profiling, exosome analysis) that will be available to the diagnostic team in the near future.
Conflict-of-interest disclosure: The author declares no competing financial interests.