Key Points
Hematopoietic cell and gene therapy can prevent in vivo infections by Mycobacteria spp. using different lentiviral vectors.
Cellular repair of macrophages in vivo highlights phagocytes as key players in the disease progression.
Abstract
Mendelian susceptibility to mycobacterial disease is a rare primary immunodeficiency characterized by severe infections caused by weakly virulent mycobacteria. Biallelic null mutations in genes encoding interferon gamma receptor 1 or 2 (IFNGR1 or IFNGR2) result in a life-threatening disease phenotype in early childhood. Recombinant interferon γ (IFN-γ) therapy is inefficient, and hematopoietic stem cell transplantation has a poor prognosis. Thus, we developed a hematopoietic stem cell (HSC) gene therapy approach using lentiviral vectors that express Ifnγr1 either constitutively or myeloid specifically. Transduction of mouse Ifnγr1−/− HSCs led to stable IFNγR1 expression on macrophages, which rescued their cellular responses to IFN-γ. As a consequence, genetically corrected HSC-derived macrophages were able to suppress T-cell activation and showed restored antimycobacterial activity against Mycobacterium avium and Mycobacterium bovis Bacille Calmette-Guérin (BCG) in vitro. Transplantation of genetically corrected HSCs into Ifnγr1−/− mice before BCG infection prevented manifestations of severe BCG disease and maintained lung and spleen organ integrity, which was accompanied by a reduced mycobacterial burden in lung and spleen and a prolonged overall survival in animals that received a transplant. In summary, we demonstrate an HSC-based gene therapy approach for IFNγR1 deficiency, which protects mice from severe mycobacterial infections, thereby laying the foundation for a new therapeutic intervention in corresponding human patients.
Introduction
Hematopoietic stem cell gene therapy (HSCGT) is a promising approach for the treatment of a growing number of primary immunodeficiencies. Numerous clinical trials are currently underway to improve the quality of life for patients suffering from severe combined immunodeficiency-X1 (SCID-X1) or adenosine deaminase SCID (ADA-SCID), Wiskott-Aldrich syndrome, or chronic granulomatous disease.1-4 Improvements in vector design and the use of safety-improved self-inactivating (SIN) lentiviral vectors have laid the foundation for today’s new generation of hematopoietic stem cell (HSC)–based gene therapy.5 Given the aforementioned success, HSCGT may be extended to hematopoietic diseases in which the impaired blood and tissue leukocytes are unable to fight mycobacterial infections.
Along this line, biallelic mutations in IFNGR1 have been associated from 1996 onward with a selective predisposition to severe infections by weakly virulent mycobacteria, including Bacille Calmette-Guérin (BCG) vaccines and environmental mycobacteria. This condition is known as Mendelian susceptibility to mycobacterial disease (MSMD) (Online Mendelian Inheritance in Man [OMIM] #209950).6,7 Mutations in several genes involved in the cross-talk between the myeloid and lymphoid cells that orchestrate interferon-γ (IFN-γ)–mediated immunity have been described in patients with MSMD. As a consequence, tissue macrophages cannot destroy mycobacteria normally. MSMD-causing genes control either the production of IFN-γ (IL12B, IL12RB1, TYK2, IRF8, ISG15, NEMO) or the response to IFN-γ (IFNGR1, IFNGR2, STAT1, IRF8, CYBB).8 The clinical features depend on the genotype.
The onset of MSMD in patients with the 2 most severe forms, autosomal recessive (AR) complete IFNγR1 or IFNγR2 deficiency, is typically before 3 years of age and is characterized by disseminated and persistent or recurrent infections by 1 or more mycobacterial species (eg, BCG, Mycobacterium chelonae, or M avium), which are life-threatening.8-13 Conservative treatment options for all patients primarily rely on vigorous treatment of mycobacterial infections with antibiotics. IFN-γ substitution therapy can restore macrophage function only in patients whose cellular responses to IFN-γ are intact or impaired but not abolished. Indeed, patients suffering from AR complete IFNγR1 (OMIM #209950) or IFNγR2 (OMIM #107470) deficiency do not respond to IFN-γ therapy.8 Together, these 2 defects have a prevalence of around 1/100 000 births worldwide and have been found in most ethnicities. At this time, 36 patients from 30 families have been described in the literature, but it is likely that many more have been diagnosed.8,14-19
In patients suffering from complete IFNγR1 or IFNγR2 deficiency, antibiotics cannot be discontinued, and allogeneic hematopoietic stem cell transplantation (allo-HSCT) is the only curative treatment.20-24 However, substantial adverse effects, such as recurrent infections during the transplantation scenario or even graft rejection, still considerably limit the success of allo-HSCT to treat AR complete IFNγR1 or IFNγR2 deficiency. An unusually high graft rejection rate is associated with high IFN-γ serum levels present in IFNγR–deficient MSMD patients because their cells are not able to clear IFN-γ from the blood.25 This negative effect can be exacerbated even if IFN-γ is produced by alloreactive T cells in the donor graft.26 As a consequence, IFN-γ may interfere with the cell cycles of donor HSCs or induce cell death by upregulating Fas ligand in combination with tumor necrosis factor-α.26-30 With 17 reported deaths in 36 patients, the overall prognosis of HSCT for patients suffering from IFNγR1 or IFNγR2 deficiency is poor, which highlights the need for improved therapeutic approaches.
Because current forms of treatment are limited, we aimed to establish an HSCGT approach to restoring the antimycobacterial function in hematopoietic cells in vivo. This approach would prevent the risks of graft rejection and graft-versus-host disease that are inherent in HSCT. To achieve this aim, we used lentiviral vector technology to correct the cellular response in the hematopoietic system to IFN-γ and investigated the ability of genetically corrected macrophages to clear mycobacteria in vitro and in vivo.
Our findings suggest that restored expression of IFNγR1 on macrophages is directly linked to the cellular repair of these cells and highlight a new cell-based treatment approach for the clinical condition of MSMD.
Methods
Design of lentiviral vectors
Ifnγr1 complementary DNA (cDNA) was introduced into third-generation SIN lentiviral vectors downstream of a spleen focus-forming virus (SFFV) promoter (Lv.SFFV.Ifnγr1), a microRNA 223 (miR223) promoter (Lv.miR223.Ifnγr1), or an SP146.gp91 synthetic myeloid-specific promoter (MSP) (Lv.MSP.Ifnγr1), and all constructs were also coupled with a green fluorescent protein (GFP) through an internal ribosome entry site.
Cultivation of cells
All cell lines and primary cells were kept under standard culture conditions at 37°C, 5% CO2, and 95% humidity. RAW264.7 cells were kept in 6-well adherent plates. Cells were kept in high glucose Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Biochrom) and 1% penicillin/streptomycin (Life Technologies).
Mice
B6.SJL-Ptprca-Pep3b/BoyJZtm (Ly5.1; CD45.1) mice were used as wild-type (WT) controls, and B6.129S7-Ifngr1tm1Agt/J (Ifnγr1−/−) mice harboring a knockout in the Ifnγr1 gene served as a disease model of MSMD. Mice were housed in the central animal facility of Hannover Medical School under specific-pathogen-free conditions in individually ventilated cages. Mice had access to food and water ad libitum. All animal experiments were approved by the Lower Saxony State Animal Welfare Committee and were conducted according to the animal welfare law.
Isolation, cultivation, and transduction of lineage-negative cells
Lineage-negative cells were isolated from total bone marrow by using a lineage cell depletion kit (Miltenyi Biotec) following the manufacturer’s instructions. Prestimulation of lineage-negative cells was performed for 24 hours in STIF medium (StemSpan medium supplemented with 1 mM penicillin/streptomycin, 2 mM L-glutamine, 100 ng/mL stem cell factor, 20 ng/mL thrombopoietin, 20 ng/mL insulinlike growth factor-2, and 100 ng/mL fibroblast growth factor-2. Thereafter, transduction of lineage-negative cells was performed on RetroNectin (Takara)-coated plates using STIF medium and multiplicities of infection between 20 and 50 as previously described.31
Bone marrow transplantation
Ifnγr1−/− recipient mice were irradiated with a single dose of 9 Gy. Lineage-negative bone marrow cells from Ifnγr1−/− or WT donor mice were isolated as described above and injected intravenously into the tail vein of recipient mice 24 hours after irradiation. Alternatively, lineage-negative cells were transduced with SIN lentiviral vectors and transplanted by the same protocol. For some experiments, nontransplanted Ifnγr1−/− mice served as negative controls.
Pulmonary BCG infections
After reconstitution of the hematopoietic system (>6 weeks after transplantation), transplanted mice were infected by intratracheal application of 1 × 107 colony-forming units (CFUs) DsRed-labeled BCG-Pasteur under general anesthesia using intraperitoneal injection of ketamine/rompun.
Differentiation of lineage-negative cells
M avium
Intracellular killing of M avium by macrophages was assessed as described in Anes et al33 with or without stimulation with IFN-γ for 24 hours. Macrophages were infected for 1 hour with M avium at an optical density (OD600) of 0.1. Non-phagocytosed bacteria were removed by intense washing. At 1 and 24 hours after infection, cells were lysed by using 0.1% Triton X100 in phosphate-buffered saline), plated on Middlebrock 7H9 agar plates, and incubated at 37°C. Colonies were counted after 2 weeks.
BCG
Transduced lineage-negative cells were sorted and differentiated into macrophages, and 200 000 cells per condition were seeded. At 24 hours before infection, the cells were washed twice with phosphate-buffered saline to remove remaining antibiotics. Cells were stimulated with 50 ng/mL IFN-γ in RPMI 1640 (Gibco) supplemented with 10% FBS. Cells were infected with BCG at an multiplicity of infection of 100 and incubated for 1 hour to allow BCG uptake. Afterward, 10 μg/mL gentamycin was added to remove all extracellular BCG. Cells were harvested and lysed at 1 hour and 24 hours after infection by using a 24-gauge cannula to resuspend the cells 10 times. Lysates were plated at different dilutions on 7H10 plates and incubated for 2 weeks until the colonies were counted. CFU per mL values were then calculated.
IFN-γ serum levels
Blood of mice was drawn from the retrobulbar vein plexus, and cell-free serum was collected by centrifugation in serum separation tubes. Serum IFN-γ levels were determined by using the eBioscience mouse IFN-γ Ready-SET-Go! enzyme-linked immunosorbent assay kit.
Light sheet microscopy
Light sheet microscopy was performed as described before.34
Histology
For histopathologic investigation, formalin-fixed paraffin-embedded lung tissue was sectioned at 2 to 3 µm thickness, mounted on glass slides, and stained with hematoxylin and eosin. For immunohistochemistry, a polyclonal rabbit anti-eGFP antibody (1:10 000; catalog No. orb303312; Biorbyt) was used. A microwave pretreatment over 20 minutes with citrate buffer was applied for antigen retrieval. The primary antibody was replaced by rabbit serum as a negative control. A peroxidase-conjugated avidin-biotin complex (Vector Laboratories) and 3.3-diaminobenzidine-tetrachloride in 0.1 M imidazole (Sigma-Aldrich) was used for visualization. Finally, sections were counterstained with Mayer’s hematoxylin (Merck).
Results
Design of lentiviral vectors and safety studies in primary hematopoietic cells
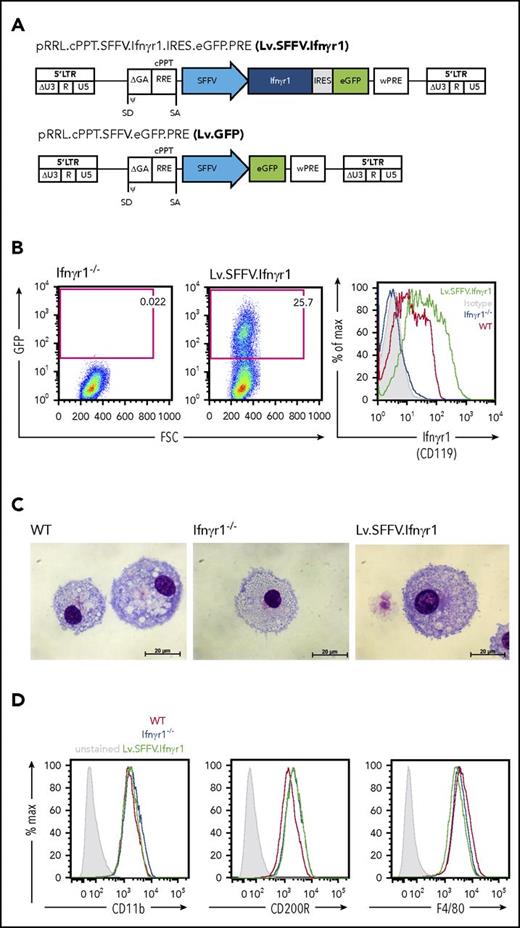
To establish an HSCGT approach for IFNγR1 deficiency, we used third-generation SIN vectors to express a murine Ifnγr1 cDNA from an SFFV virus promoter/enhancer element. To assess transgene expression, we coupled the Ifnγr1 cDNA via an internal ribosomal entry site to an enhanced GFP (Lv.SFFV.Ifnγr1). The same lentiviral vector backbone was used to establish the control vector, which expressed the GFP transgene directly under the control of the SFFV promoter (Lv.SFFV.GFP) (Figure 1A). As a first step, we evaluated vector-mediated transgene expression in the murine fibroblast cell line SC1, resulting in detectable levels of IFNγR1 (CD119) in cell lysates and on the cell surface (supplemental Figure 1A-C, available on the Blood Web site). Transgene expression was next evaluated in lineage-negative hematopoietic stem/progenitor cells (HSPCs) of mice deficient in Ifnγr1 (Ifnγr1−/−) and which faithfully recapitulate the MSMD phenotype upon infection with BCG.25,35 Transduction of lineage-negative HSPCs revealed co-expression of GFP and IFNγR1 in clear contrast to nontransduced Ifnγr1−/− control cells (Figure 1B). Of note, mean fluorescent intensity values of CD119 indicated higher expression levels of IFNγR1 in transduced Ifnγr1−/− cells compared with their WT counterparts. As a next step, we followed a straightforward differentiation protocol using macrophage colony-stimulating factor to differentiate HSPCs toward macrophages. After 7 days of differentiation, macrophages derived from Lv.SFFV.Ifnγr1-transduced Ifnγr1−/− HSPCs showed typical morphology on cytospins and surface marker expression of CD11b, CD200R, and F4/80, which was comparable to that in HSPC-derived macrophages from WT or noncorrected Ifnγr1−/− cells (Figure 1C-D; supplemental Figure 2). Furthermore, stable transgene expression as well as normal rates of cell death and cell proliferation were observed in Lv.SFFV.Ifnγr1-transduced Raw 264.7 cells, which highlights the functional expression of IFNγR1 upon transduction of our lentiviral vector (supplemental Figure 3A-C).
Constitutive lentiviral Ifnγr1 overexpression in Ifnγr1−/−macrophages. (A) Schematic representation of the lentiviral vector Lv.SFFV.Ifnγr1 expressing the Ifnγr1 cDNA by an SFFV promoter coupled to a GFP reporter via an internal ribosomal entry site (IRES). (B) GFP expression in Ifnγr1−/− macrophages (left) without and (middle) with transduction using the Lv.SFFV.Ifnγr1 vector. (Right) Histogram depicting IFNγR1 (CD119) expression in Ifnγr1−/−, WT, and Lv.SFFV.Ifnγr1-transduced macrophages compared with an isotype control. (C) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1 macrophages presenting characteristic morphology in representative cytospins. (D) Characteristic macrophage surface marker expression of CD11b, CD200R, and F4/80 at similar levels on WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages compared with unstained samples. cPPT, central polypurine tract; FSC, forward scatter; GA, truncated GAG (group-specific antigen) sequence; LTR, long terminal repeat; ψ, packaging signal; R, redundant region; RRE, Rev-responsive element; SA, splice acceptor site; SD, splice donor site; U3, U3 region; U5, U5 region; wPRE, Woodchuck hepatitis virus posttranscriptional response element; WT, wild type.
Constitutive lentiviral Ifnγr1 overexpression in Ifnγr1−/−macrophages. (A) Schematic representation of the lentiviral vector Lv.SFFV.Ifnγr1 expressing the Ifnγr1 cDNA by an SFFV promoter coupled to a GFP reporter via an internal ribosomal entry site (IRES). (B) GFP expression in Ifnγr1−/− macrophages (left) without and (middle) with transduction using the Lv.SFFV.Ifnγr1 vector. (Right) Histogram depicting IFNγR1 (CD119) expression in Ifnγr1−/−, WT, and Lv.SFFV.Ifnγr1-transduced macrophages compared with an isotype control. (C) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1 macrophages presenting characteristic morphology in representative cytospins. (D) Characteristic macrophage surface marker expression of CD11b, CD200R, and F4/80 at similar levels on WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages compared with unstained samples. cPPT, central polypurine tract; FSC, forward scatter; GA, truncated GAG (group-specific antigen) sequence; LTR, long terminal repeat; ψ, packaging signal; R, redundant region; RRE, Rev-responsive element; SA, splice acceptor site; SD, splice donor site; U3, U3 region; U5, U5 region; wPRE, Woodchuck hepatitis virus posttranscriptional response element; WT, wild type.
Phenotypic correction of IFN-γ functionality upon constitutive IFNγR1 overexpression
After we evaluated SFFV-mediated IFNγR1 expression on Ifnγr1−/− knockout and WT cells, we studied the direct effect of IFN-γ on corrected macrophages. Because expression of major histocompatibility complex class II on macrophages can be induced upon stimulation with IFN-γ,36 we studied the upregulation of HLA-DR and of the co-stimulatory molecule CD86 (B7.2) upon treatment with IFN-γ. Overexpression of IFNγR1 by the Lv.SFFV.Ifnγr1 construct was able to restore upregulation of HLA-DR in GFP+ macrophages upon stimulation with IFN-γ, whereas noncorrected Ifnγr1−/− cells showed no response (Figure 2A). A similar observation was made for CD86. Here, similar to HLA-DR, only corrected GFP+ macrophages showed restored upregulation of CD86 compared with WT cells (Figure 2B). Proper downstream signaling of IFN-γ is induced upon initial binding to IFNγR1 and subsequent dimerization of the 2 IFNγR1 chains and the assembly of the functional IFNγR1/2 heterodimer receptor complex. Although the IFN-γ:IFNγR1/2 receptor complex forms a STAT1 binding site at the intracellular domain of IFNγR1, receptor-associated activated JAK kinases are able to phosphorylate STAT1 and to induce proper downstream signaling.37 Cultivation of Lv.SFFV.Ifnγr1-transduced macrophages led to IFN-γ consumption over time, so that after 24 hours of cultivation, only 60% to 80% of the original IFN-γ input was detected (Figure 2C). A similar picture was observed for the ability to phosphorylate STAT1 in the presence of IFN-γ. Although WT or genetically corrected macrophages were able to phosphorylate STAT1, noncorrected Ifnγr1−/− macrophages showed no phosphorylated STAT1 upregulation in the presence of IFN-γ (Figure 2D). Proper downstream signaling was also evaluated for known downstream target genes of IFN-γ such as interferon regulatory factor 1 (Irf1), nitric oxide synthase 2 (Nos2), or indoleamine 2,3-dioxygenase (Ido). Stimulation of macrophages with IFN-γ provided a restored expression pattern of Irf1, Nos2, and Ido in macrophages transduced with the Lv.SFFV.Ifnγr1 construct, which was very similar to WT cells (Figure 2E). In contrast, macrophages from Ifnγr1−/− showed only background expression of the aforementioned genes (Figure 2E). We showed that surface expression of HLA-DR and the secretion of IDO from macrophages have a direct effect on T-cell behavior; thus, we also analyzed the IFN-γ–dependent ability of macrophages to suppress ovalbumin-specific T-cell proliferation in the presence or absence of antigen.38 Cultivation of CD4+ T cells from OTII mice with ovalbumin-loaded and IFN-γ–primed macrophages showed a clear reduction in the proliferation of T cells when either WT or genetically corrected macrophages were present (Figure 2F). In contrast, more than 80% of T cells still proliferated if they were cocultured with Ifnγr1−/− macrophages (Figure 2F). As a final assessment of our lentiviral construct, we elucidated the IFN-γ–dependent intracellular degradation of M avium or BCG. Irrespective of the mycobacterium used, genetically corrected macrophages showed restored intracellular degradation of either M avium or BCG, which was at a level comparable to WT macrophages (Figure 2G-H).
Restored macrophage functionality upon Lv.SFFV.Ifnγr1 transduction of Ifnγr1−/−cells. (A) Representative histograms depicting major histocompatibility class II (MHC-II) upregulation on IFN-γ-stimulated vs unstimulated (unstim) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 3 independent experiments (two-way analysis of variance [ANOVA] using Sidak’s multiple comparisons post hoc testing. (B) Representative histograms depict CD86 upregulation on IFN-γ-stimulated vs unstimulated WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 3 independent experiments (two-way ANOVA using Sidak’s multiple comparisons post hoc testing). (C) IFN-γ internalization of WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages over 24 hours (normalized to 0 hours; n = 4; two-way ANOVA using Tukey’s multiple comparisons post hoc testing). (D) Phosphorylation of STAT1 (pSTAT1) in unstimulated (–) and IFN-γ-stimulated (+) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages compared with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control. (E) Relative messenger RNA expression of Irf1, Nos2, and Ido in IFN-γ–stimulated WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages normalized to unstimulated samples (Irf1, n = 4; Nos2 and Ido, n = 3 each). (F) Induction of ovalbumin (OVA)-specific T-cell proliferation by WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages upon stimulation with IFN-γ or IFN-γ and OVA. Bars depict the percentage of eFluor670low CD4+ T cells (n = 3; two-way ANOVA using Bonferroni’s multiple comparisons post hoc testing). (G) Bacterial burden of M avium in macrophages 0 hours and 24 hours after infection (n = 3; two-way ANOVA using Sidak’s multiple comparisons post hoc testing). (H) Bacterial burden of Bacille Calmette-Guérin colonies in macrophages 8 days after infection (normalized to uptake value 4 hours after infection; n = 3; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. For all bars, mean and standard error of the mean (SEM) are shown. ns, not significant.
Restored macrophage functionality upon Lv.SFFV.Ifnγr1 transduction of Ifnγr1−/−cells. (A) Representative histograms depicting major histocompatibility class II (MHC-II) upregulation on IFN-γ-stimulated vs unstimulated (unstim) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 3 independent experiments (two-way analysis of variance [ANOVA] using Sidak’s multiple comparisons post hoc testing. (B) Representative histograms depict CD86 upregulation on IFN-γ-stimulated vs unstimulated WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 3 independent experiments (two-way ANOVA using Sidak’s multiple comparisons post hoc testing). (C) IFN-γ internalization of WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages over 24 hours (normalized to 0 hours; n = 4; two-way ANOVA using Tukey’s multiple comparisons post hoc testing). (D) Phosphorylation of STAT1 (pSTAT1) in unstimulated (–) and IFN-γ-stimulated (+) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages compared with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control. (E) Relative messenger RNA expression of Irf1, Nos2, and Ido in IFN-γ–stimulated WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages normalized to unstimulated samples (Irf1, n = 4; Nos2 and Ido, n = 3 each). (F) Induction of ovalbumin (OVA)-specific T-cell proliferation by WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages upon stimulation with IFN-γ or IFN-γ and OVA. Bars depict the percentage of eFluor670low CD4+ T cells (n = 3; two-way ANOVA using Bonferroni’s multiple comparisons post hoc testing). (G) Bacterial burden of M avium in macrophages 0 hours and 24 hours after infection (n = 3; two-way ANOVA using Sidak’s multiple comparisons post hoc testing). (H) Bacterial burden of Bacille Calmette-Guérin colonies in macrophages 8 days after infection (normalized to uptake value 4 hours after infection; n = 3; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. For all bars, mean and standard error of the mean (SEM) are shown. ns, not significant.
Lineage restrictive expression of Ifnγr1 mediates phenotypic correction in HSPC-derived macrophages
After achieving promising in vitro results using the constitutive Lv.SFFV.Ifnγr1 lentiviral construct, we designed third-generation SIN lentiviral constructs that are able to express the murine Ifnγr1 cDNA in a cell type–specific manner in cells of the myeloid lineage. To accomplish this, we used either a fragment of the human miRNA223 promoter39 (Lv.miR223.Ifnγr1) or a promoter harboring a 1.5-kb minimal promoter sequence from the gp91phox locus fused to the SP146 MSP (Lv.MSP.Ifnγr1) (Figure 3A), previously shown to express transgenes in the myeloid lineage.40,41 To evaluate the aforementioned constructs, we again isolated HSPCs from Ifnγr1−/− mice, transduced them, and differentiated the cells toward macrophages. Irrespective of the constructs used, mean fluorescent intensity values of transduced macrophages revealed distinct expression of IFNγR1, which was significantly higher than that in nontransduced Ifnγr1−/− macrophages (Figure 3B-C). Of note, expression mediated by the Lv.MSP.Ifnγr1 construct revealed the highest expression of IFNγR1 compared with WT and Lv.miR223.Ifnγr1-transduced macrophages (Figure 3B-C). After the detection of CD119 on the cell surface, we also evaluated key macrophage functions, which are dependent on IFN-γ stimulation. Similar to studies performed with the Lv.SFFV.Ifnγr1 construct, myeloid-specific expression of IFNγR1 was able to restore the upregulation of HLA-DR and CD86 in GFP+ macrophages (Figure 3D-E). In addition, both Lv.miR223.Ifnγr1 and Lv.MSP.Ifnγr1 gene-corrected cells were able to restore the expression of the IFNγR downstream targets Irf1 and Nos2 after stimulation of macrophages with IFN-γ (Figure 3F). As a final assessment for our myeloid cell–specific gene therapy approach, we also tested the ability of genetically corrected cells to reduce IFN-γ levels in cell culture supernatant and to clear intracellular BCG. HSPCs transduced with either Lv.miR223.Ifnγr1 or Lv.MSP.Ifnγr1 constructs and differentiated toward macrophages showed restored consumption of IFN-γ from the medium, reaching levels of 40% reduction within 32 hours after treatment, whereas IFN-γ levels in cultures with noncorrected Ifnγr1−/− macrophages remained unaffected (Figure 3G). Similarly, genetically corrected GFP+ macrophages showed restored intracellular degradation of BCG within 24 hours after infection, whereas GFP– cells from the same transduction were not able to clear BCG (Figure 3H). Similar to Lv.SFFV.Ifnγr1-transduced cells, Lv.miR223.Ifnγr1- or Lv.MSP.Ifnγr1-transduced RAW264.7 cells showed stable transgene expression, which was in accordance with normal rates of apoptosis and proliferation (supplemental Figure 3A-C).
Expression and function of myeloid-specific lentiviral Ifnγr1 overexpression in Ifnγr1−/−macrophages. (A) Schematic representation of the lentiviral vectors expressing Ifnγr1 cDNA by a synthetic MSP (Lv.MSP.Ifnγr1) or an miRNA223 promoter (Lv.miR223.Ifnγr1), both coupled to a GFP reporter via an IRES. (B) Histogram depicting IFNγR1 expression in Ifnγr1−/−, WT, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages. Bar graphs depict the mean fluorescence intensity (MFI) of IFNγR1 (CD119; n = 3; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (C) Representative brightfield, GFP–, and autofluorescence photos of mock-transduced, Lv.miR223.Ifnγr1-transduced, and Lv.MSP.Ifnγr1-transduced macrophages. Scale bars represent 50 μm. (D) Representative histograms depicting MHC-II upregulation on IFN-γ–stimulated vs unstimulated WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 4 independent experiments (one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (E) Representative histograms depicting CD86 upregulation on IFN-γ–stimulated vs unstimulated WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 4 independent experiments (one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (F) Relative mRNA expression of Irf1 and Nos2 in IFN-γ–stimulated WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages normalized to unstimulated samples (n = 4). (G) IFN-γ internalization of WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages over 32 hours (normalized to 0 hours; n = 3; two-way ANOVA using Tukey’s multiple comparisons post hoc testing). (H) Bacterial burden of BCG in Lv.MSP.Ifnγr1-transduced and Lv.miR223.Ifnγr1-transduced macrophages 24 hours after infection sorted into positive and negative groups for GFP expression (normalized to uptake value 0 hours after infection; n = 3). *P ≤ .05; **P ≤ .01; ****P ≤ .0001. For all bars, mean and SEM are shown.
Expression and function of myeloid-specific lentiviral Ifnγr1 overexpression in Ifnγr1−/−macrophages. (A) Schematic representation of the lentiviral vectors expressing Ifnγr1 cDNA by a synthetic MSP (Lv.MSP.Ifnγr1) or an miRNA223 promoter (Lv.miR223.Ifnγr1), both coupled to a GFP reporter via an IRES. (B) Histogram depicting IFNγR1 expression in Ifnγr1−/−, WT, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages. Bar graphs depict the mean fluorescence intensity (MFI) of IFNγR1 (CD119; n = 3; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (C) Representative brightfield, GFP–, and autofluorescence photos of mock-transduced, Lv.miR223.Ifnγr1-transduced, and Lv.MSP.Ifnγr1-transduced macrophages. Scale bars represent 50 μm. (D) Representative histograms depicting MHC-II upregulation on IFN-γ–stimulated vs unstimulated WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 4 independent experiments (one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (E) Representative histograms depicting CD86 upregulation on IFN-γ–stimulated vs unstimulated WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 4 independent experiments (one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (F) Relative mRNA expression of Irf1 and Nos2 in IFN-γ–stimulated WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages normalized to unstimulated samples (n = 4). (G) IFN-γ internalization of WT, Ifnγr1−/−, Lv.MSP.Ifnγr1-transduced, and Lv.miR223.Ifnγr1-transduced macrophages over 32 hours (normalized to 0 hours; n = 3; two-way ANOVA using Tukey’s multiple comparisons post hoc testing). (H) Bacterial burden of BCG in Lv.MSP.Ifnγr1-transduced and Lv.miR223.Ifnγr1-transduced macrophages 24 hours after infection sorted into positive and negative groups for GFP expression (normalized to uptake value 0 hours after infection; n = 3). *P ≤ .05; **P ≤ .01; ****P ≤ .0001. For all bars, mean and SEM are shown.
Hematopoietic gene and cell therapy using genetically corrected HSPCs mediates protection against BCG infection in vivo
As an in vivo assessment of our gene therapy approach, we used Ifnγr1−/− mice and established an HSCGT approach, with the overall aim of protecting mice that had received transplants from BCG infection (Figure 4A). After 9 Gy lethal irradiation, Ifnγr1−/− recipient mice received lineage-negative cells from either WT C57BL/6 or noncorrected Ifnγr1−/− donor mice. Alternatively, Ifnγr1−/− recipient mice were transplanted with genetically corrected lineage-negative cells transduced with either Lv.SFFV.Ifnγr1 or Lv.MSP.Ifnγr1 constructs (Figure 4A). After a minimum time of 6 weeks to allow for reconstitution of the hematopoietic system, we performed intrapulmonary infection of transplanted mice with DsRed-BCG42 followed by observation for 5 to 9 weeks (Figure 4A). Analyzing the vector copy numbers (VCNs) in the bone marrow of transplanted animals, mice receiving Lv.GFP-transduced cells showed a VCN of 8 to 11, whereas cells from animals transplanted with Lv.SFFV.Ifnγr1 or Lv.MSP.Ifnγr1 had a VCN of 1 to 2 or 0.5 to 2, respectively (Figure 4B). Of note, VCN values in the lungs increased after BCG infection in animals that received genetically corrected cells whereas mice transplanted with Lv.GFP-transduced HSCs showed stable VCN values (Figure 4B). Detection of genetically corrected cells in the bone marrow and lung of transplanted mice was associated with stabilized body weight ratios and with an increase in overall survival after BCG infection (Figure 4C; supplemental Figure 4A). Irrespective of the viral construct used, Ifnγr1−/− mice transplanted with genetically corrected cells survived more than 2 months after infection, whereas control mice showed severe BCG-related symptoms at 21 days after infection (Figure 4C). Similarly, mice that received genetically corrected cells showed low levels of IFN-γ throughout the infection period, whereas Lv.GFP-transplanted mice showed increased levels of IFN-γ (Figure 4D).
Engraftment of lentiviral Ifnγr1 overexpressing cells rescues Ifnγr1−/−mice from lethal pulmonary BCG infection. (A) Schematic representation of the experimental outline. Ifnγr1−/− mice were lethally irradiated and transplanted with lineage-negative Ifnγr1−/− cells transduced with Lv.SFFV.Ifnγr1 or Lv.MSP.Ifnγr1. After hematopoietic reconstitution, DsRed-labeled BCG was instilled into the lungs of the transplanted mice. Final analysis was performed 9 weeks after infection. (B) VCNs integrated into cells in the bone marrow (BM) and lungs (L) of transplanted and infected mice. Equal-size symbols represent VCNs of the same mouse in BM and L (n ≥ 3). (C) Kaplan-Meier curve depicting the survival of mice after BCG infection in 2 independent experiments (positive control [ctrl], 6 mice receiving WT cells; negative control, 13 mice receiving Lv.GFP-transduced Ifnγr1−/− cells, 7 mice receiving Lv.SFFV.Ifnγr1, and 6 mice receiving Lv.MSP.Ifnγr1. (D) IFN-γ serum levels of mice at 2, 4, 6, and 8 weeks after infection. Mean and SEM are shown for 3 independent experiments (positive control, 9 mice that received WT cells; negative control, 9 mice that received Lv.GFP-transduced Ifnγr1−/− cells and 7 nontransplanted Ifnγr1−/− mice that received Lv.SFFV.Ifnγr1 and Lv.MSP.Ifnγr1.
Engraftment of lentiviral Ifnγr1 overexpressing cells rescues Ifnγr1−/−mice from lethal pulmonary BCG infection. (A) Schematic representation of the experimental outline. Ifnγr1−/− mice were lethally irradiated and transplanted with lineage-negative Ifnγr1−/− cells transduced with Lv.SFFV.Ifnγr1 or Lv.MSP.Ifnγr1. After hematopoietic reconstitution, DsRed-labeled BCG was instilled into the lungs of the transplanted mice. Final analysis was performed 9 weeks after infection. (B) VCNs integrated into cells in the bone marrow (BM) and lungs (L) of transplanted and infected mice. Equal-size symbols represent VCNs of the same mouse in BM and L (n ≥ 3). (C) Kaplan-Meier curve depicting the survival of mice after BCG infection in 2 independent experiments (positive control [ctrl], 6 mice receiving WT cells; negative control, 13 mice receiving Lv.GFP-transduced Ifnγr1−/− cells, 7 mice receiving Lv.SFFV.Ifnγr1, and 6 mice receiving Lv.MSP.Ifnγr1. (D) IFN-γ serum levels of mice at 2, 4, 6, and 8 weeks after infection. Mean and SEM are shown for 3 independent experiments (positive control, 9 mice that received WT cells; negative control, 9 mice that received Lv.GFP-transduced Ifnγr1−/− cells and 7 nontransplanted Ifnγr1−/− mice that received Lv.SFFV.Ifnγr1 and Lv.MSP.Ifnγr1.
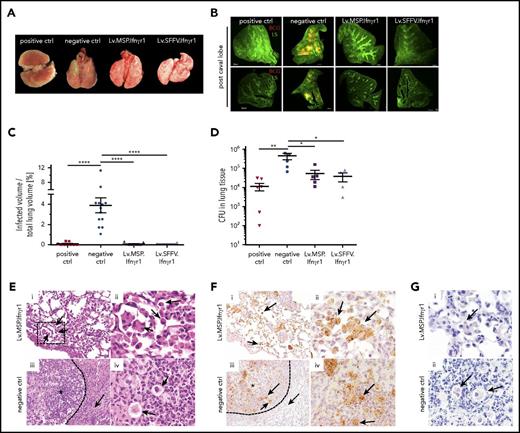
Hematopoietic gene and cell therapy sustains organ integrity of Ifnγr1−/− mice after BCG infection
We next performed detailed analysis of hematopoietic organs after immunization with BCG. Transplantation of Lv.SFFV.Ifnγr1- or Lv.MSP.Ifnγr1-transduced cells into Ifnγr1−/− mice prevented splenomegaly and led to a normalized spleen morphology and spleen-to-body-weight ratio (Figure 5A-B). In addition, mice receiving genetically corrected cells had a splenic parenchyma with characteristic regions described as red and white pulp (Figure 5C). Moreover, detailed histology of spleen sections receiving genetically corrected cells revealed only mild multifocal granulomatous inflammation irrespective of the lentiviral construct used. In contrast, negative control mice showed moderate to severe multifocal granulomatous inflammation with multinucleated giant cells. In line with the aforementioned histopathology, mice that received genetically corrected cells showed a 10- to 100-fold decrease in the mycobacterial burden in spleen homogenates compared with negative control mice (Figure 5D). A similar protective effect was observed in the lung. Ifnγr1−/− mice that received Lv.SFFV.Ifnγr1- or Lv.MSP.Ifnγr1-transduced cells had a normal lung morphology comparable to that of mice receiving healthy WT cells (positive controls), whereas mice that received noncorrected cells (negative control) showed clear signs of BCG-mediated infection (Figure 6A). The severity of BCG infection was also seen in light sheet microscopy analysis as DsRed-positive BCG infections in the post-caval lung lobe of negative control mice (Figure 6A-B). In contrast, mice receiving healthy or genetically corrected HSCs had a much lower DsRed signal, indicating a decreased BCG burden in the lung (Figure 6A-B; supplemental Videos 1-4). Similar observations were made for the inferior, middle, left, and superior lung lobes (supplemental Figure 4B). Furthermore, total CFU analysis of lung tissue confirmed the reduced mycobacterial burden in the lungs of transplanted mice. Although negative control mice had a mean of 3 × 105 CFUs per lung, mice that received genetically corrected cells showed bacterial burdens reduced by an order of magnitude (∼3 × 104 CFUs per lung), irrespective of the lentiviral construct used (Figure 6D). Lung histology further supported the protective effect of genetically corrected cells in the lung. Although animals transplanted with Lv.MSP.Ifnγr1 showed a normal lung structure (Figure 6Ei-ii) and the presence of GFP-labeled macrophages in the bronchoalveolar space (Figure 6Fi-ii), Lv.GFP-transplanted negative control mice showed formation of pulmonary granulomas (indicated by an asterisk) with multinucleated giant cells surrounded by collagen fibers (Figure 6Eiii-iv and Fiii-iv). Although Ziehl-Neelsen staining of macrophages in mice transplanted with Lv.MSP.Ifnγr1 or Lv.GFP revealed the presence of acid-fast bacteria (Figure 6G), only macrophages from mice transplanted with Lv.MSP.Ifnγr1-transduced cells were able to degrade BCG whereas Lv.GFP macrophages started to form granulomas (Figure 6D-G).
Prevention of severe splenomegaly upon constitutive or myeloid-specific Ifnγr1 overexpression. (A) Representative macroscopic photos of spleens isolated from mice 9 weeks after infection. Positive control, mouse that received WT cells; negative control, nontransplanted Ifnγr1−/− mouse. (B) Spleen-to-body weight ratio depicting the normalized spleen weight of individual mice 9 weeks after infection. Mean and SEM are shown for 3 independent experiments (positive control, 10 mice that received WT cells; negative control, 13 mice that received Lv.GFP-transduced Ifnγr1−/− cells and nontransplanted Ifnγr1−/− mice; 10 mice received Lv.SFFV.Ifnγr1; 9 mice received Lv.MSP.Ifnγr1; one-way ANOVA using Tukey’s multiple comparisons post hoc testing). (C) Hematoxylin and eosin–stained histologic sections of spleens isolated 9 weeks after infection. Positive controls (mice that received WT cells), Lv.SFFV.Ifnγr1 mice, and Lv.MSP.Ifnγr1 mice showed typical spleen histology with clear separation between white pulp (areas within dotted lines [#]) and red pulp (area indicated by an asterisk), whereas this separation is absent in negative control mice (those that received Lv.GFP-transduced Ifnγr1−/− cells). Original magnification ×100. (D) BCG CFU counts of spleen homogenates 9 weeks after infection. Mean and SEM are shown for 2 independent experiments (positive control, 7 mice that received WT cells; negative control, 6 mice that received Lv.GFP-transduced Ifnγr1−/− cells; nontransplanted Ifnγr1−/− mice: Lv.SFFV.Ifnγr1, n = 4; Lv.MSP.Ifnγr1, n = 5). ***P ≤ .001.
Prevention of severe splenomegaly upon constitutive or myeloid-specific Ifnγr1 overexpression. (A) Representative macroscopic photos of spleens isolated from mice 9 weeks after infection. Positive control, mouse that received WT cells; negative control, nontransplanted Ifnγr1−/− mouse. (B) Spleen-to-body weight ratio depicting the normalized spleen weight of individual mice 9 weeks after infection. Mean and SEM are shown for 3 independent experiments (positive control, 10 mice that received WT cells; negative control, 13 mice that received Lv.GFP-transduced Ifnγr1−/− cells and nontransplanted Ifnγr1−/− mice; 10 mice received Lv.SFFV.Ifnγr1; 9 mice received Lv.MSP.Ifnγr1; one-way ANOVA using Tukey’s multiple comparisons post hoc testing). (C) Hematoxylin and eosin–stained histologic sections of spleens isolated 9 weeks after infection. Positive controls (mice that received WT cells), Lv.SFFV.Ifnγr1 mice, and Lv.MSP.Ifnγr1 mice showed typical spleen histology with clear separation between white pulp (areas within dotted lines [#]) and red pulp (area indicated by an asterisk), whereas this separation is absent in negative control mice (those that received Lv.GFP-transduced Ifnγr1−/− cells). Original magnification ×100. (D) BCG CFU counts of spleen homogenates 9 weeks after infection. Mean and SEM are shown for 2 independent experiments (positive control, 7 mice that received WT cells; negative control, 6 mice that received Lv.GFP-transduced Ifnγr1−/− cells; nontransplanted Ifnγr1−/− mice: Lv.SFFV.Ifnγr1, n = 4; Lv.MSP.Ifnγr1, n = 5). ***P ≤ .001.
Preservation of lung structure after pulmonary BCG infection upon constitutive or myeloid-specific Ifnγr1 overexpression. (A) Representative macroscopic photos of lungs isolated from mice 9 weeks after infection (positive control, mouse that received WT cells; negative control, mouse that received Lv.GFP-transduced Ifnγr1−/− cells). (B) Representative light sheet microscopy photos of post caval lung lobes depicting the lung structure (LS; green) and infiltration of DsRed-labeled BCG (red) (positive control, mouse that received WT cells; negative control, nontransplanted Ifnγr1−/− mouse). Scale bars represent 500 μm. (C) Quantification of light sheet microscopy depicting the percentage of infected volume of the total lung volume. Mean and SEM are shown for 1 representative experiment (positive control, 9 mice that received WT cells; negative control, 13 nontransplanted Ifnγr1−/− mice; Lv.SFFV.Ifnγr1, n = 10; Lv.MSP.Ifnγr1, n = 9; one-way ANOVA using Tukey’s multiple comparisons post hoc testing). (D) BCG CFU counts of lung homogenates 9 weeks after infection. Mean and SEM (error bars) are shown for 2 independent experiments (positive control, 6 mice that received WT cells; negative control, 5 mice that received Lv.GFP-transduced Ifnγr1−/− cells and nontransplanted Ifnγr1−/− mice: Lv.SFFV.Ifnγr1, n = 4; Lv.MSP.Ifnγr1, n = 5; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (E) Representative hematoxylin and eosin–stained histologic sections of lungs isolated 9 weeks after infection are shown for (i-ii) Lv.MSP.Ifnγr1 mouse and (iii-iv) negative control (mouse that received Lv.GFP-transduced Ifnγr1−/− cells). (i) Macrophages and multinucleated cells within the alveoli indicated by black arrows (original magnification ×200). (ii) Higher magnification of black box indicated in (i). (iii) Granuloma indicated by asterisk and macrophages and neutrophils surrounded by collagen fibers (dotted arrow) (original magnification ×200). (iv) Higher magnification photo of giant cells (black arrows). (F) Representative GFP-stained histologic lung sections isolated 9 weeks after infection are shown for (i-ii) Lv.MSP.Ifnγr1 mouse and (iii-iv) negative control mouse that received Lv.GFP-transduced Ifnγr1−/− cells. (i) GFP+ cells (brown signals) are distributed equally throughout Lv.MSP.Ifnγr1 mouse lung tissue and give rise to macrophages (black arrows; ×200). (ii) Higher magnification (×600) of GFP+ intra-alveolar macrophages. (iii) GFP+ cells are numerous in granulomas of negative control mice (*) and macrophages and neutrophils (black arrows) surrounded by collagen fibers (dotted arrow; ×200). (iv) Higher magnification (×600) photo of clustered GFP+ cells (black arrows). (G) Ziehl-Neelson staining of histologic lung sections indicating intracellular acid-fast bacteria (black arrows) (original magnification ×600 for i-ii). *P ≤ .05; **P ≤ .01; ****P ≤ .0001.
Preservation of lung structure after pulmonary BCG infection upon constitutive or myeloid-specific Ifnγr1 overexpression. (A) Representative macroscopic photos of lungs isolated from mice 9 weeks after infection (positive control, mouse that received WT cells; negative control, mouse that received Lv.GFP-transduced Ifnγr1−/− cells). (B) Representative light sheet microscopy photos of post caval lung lobes depicting the lung structure (LS; green) and infiltration of DsRed-labeled BCG (red) (positive control, mouse that received WT cells; negative control, nontransplanted Ifnγr1−/− mouse). Scale bars represent 500 μm. (C) Quantification of light sheet microscopy depicting the percentage of infected volume of the total lung volume. Mean and SEM are shown for 1 representative experiment (positive control, 9 mice that received WT cells; negative control, 13 nontransplanted Ifnγr1−/− mice; Lv.SFFV.Ifnγr1, n = 10; Lv.MSP.Ifnγr1, n = 9; one-way ANOVA using Tukey’s multiple comparisons post hoc testing). (D) BCG CFU counts of lung homogenates 9 weeks after infection. Mean and SEM (error bars) are shown for 2 independent experiments (positive control, 6 mice that received WT cells; negative control, 5 mice that received Lv.GFP-transduced Ifnγr1−/− cells and nontransplanted Ifnγr1−/− mice: Lv.SFFV.Ifnγr1, n = 4; Lv.MSP.Ifnγr1, n = 5; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). (E) Representative hematoxylin and eosin–stained histologic sections of lungs isolated 9 weeks after infection are shown for (i-ii) Lv.MSP.Ifnγr1 mouse and (iii-iv) negative control (mouse that received Lv.GFP-transduced Ifnγr1−/− cells). (i) Macrophages and multinucleated cells within the alveoli indicated by black arrows (original magnification ×200). (ii) Higher magnification of black box indicated in (i). (iii) Granuloma indicated by asterisk and macrophages and neutrophils surrounded by collagen fibers (dotted arrow) (original magnification ×200). (iv) Higher magnification photo of giant cells (black arrows). (F) Representative GFP-stained histologic lung sections isolated 9 weeks after infection are shown for (i-ii) Lv.MSP.Ifnγr1 mouse and (iii-iv) negative control mouse that received Lv.GFP-transduced Ifnγr1−/− cells. (i) GFP+ cells (brown signals) are distributed equally throughout Lv.MSP.Ifnγr1 mouse lung tissue and give rise to macrophages (black arrows; ×200). (ii) Higher magnification (×600) of GFP+ intra-alveolar macrophages. (iii) GFP+ cells are numerous in granulomas of negative control mice (*) and macrophages and neutrophils (black arrows) surrounded by collagen fibers (dotted arrow; ×200). (iv) Higher magnification (×600) photo of clustered GFP+ cells (black arrows). (G) Ziehl-Neelson staining of histologic lung sections indicating intracellular acid-fast bacteria (black arrows) (original magnification ×600 for i-ii). *P ≤ .05; **P ≤ .01; ****P ≤ .0001.
Discussion
In this study, we demonstrated that an HSCGT approach to treat IFNγR1 deficiency is feasible and that lentiviral gene transfer can correct the IFN-γ signaling pathway in HSC-derived macrophages in vitro. Thus, we established an HSCGT approach using Ifnγr1−/− mice and evaluated the therapeutic benefit of our HSCGT approach in a clinically relevant infection model using BCG. Thus, we highlight a new treatment approach, which is able to restore antimycobacterial activity of macrophages in lung and spleen. This observation is of clinical relevance, because patients suffering from AR complete IFNγR1 deficiency have poor prognosis and often die in early childhood as a result of disseminated infections with BCG and/or other environmental mycobacteria.8,10
The feasibility and clinical transfer of HSCGT using retroviral vector technology has already been proven for other primary immunodeficiencies such as Wiskott-Aldrich syndrome, chronic granulomatous disease, SCID-X1, or ADA-SCID,1-4 leading to the first marketing approval of an ex vivo gene therapy.43 In our model, which is similar to the aforementioned approaches, we have transplanted genetically corrected HSCs into lethally irradiated Ifnγr1−/− mice before BCG infection. In this transplantation scenario, HSCs are able to migrate to the bone marrow and engraft in the host. However, this approach might not be feasible in a transplantation scenario in which genetically corrected HSCs will be transplanted into patients who carry disseminated mycobacterial infections. Here, high levels of IFN-γ in the plasma of patients may interfere with the engraftment of donor HSCs. As with similar attempts to use HSCT with HLA-identical relatives, IFN-γ may induce proliferation of genetically corrected HSCs, which hampers the engraftment potential of the HSCs and could ultimately lead to graft rejection.22,27 To circumvent this problem, humanized anti-IFN-γ antibody (fontolizumab [HuZAF]) could be used in combination with rigorous antibiotic treatment to open a therapeutic window for the transplantation of autologous and genetically corrected HSCs.44
As an alternative to antibiotic/HuZAF therapy, systemic transplantation of genetically corrected or healthy macrophages could be performed in AR complete IFNγR1– or IFNγR2–deficient patients. Here, transplanted macrophages may be able to eradicate disseminated infections and to clear IFN-γ from the plasma at the same time, thus opening a therapeutic window for subsequent HSCGT. To strengthen this innovative approach, the intrapulmonary transplantation of macrophages was shown to be very effective for the treatment of hereditary pulmonary alveolar proteinosis (PAP).45,46 Macrophages transferred directly into the lungs of diseased mice with PAP could engraft and improve PAP-related disease symptoms in a very short time frame, which makes this concept potentially suitable as a bridging therapy for AR complete IFNγR1 or IFNγR2 deficiencies. In our study, we performed HSCGT using genetically corrected HSCs. The therapeutic benefit observed in vivo, however, is presumably based on corrected macrophages, which are derived from the donor graft. In fact, a direct link between the pathophysiology of MSMD and macrophages remains elusive. To shed light on the contribution of myeloid cells to the disease progression of MSMD, we used lentiviral vectors, which express the therapeutic Ifnγr1 transgene in a myeloid cell type–specific fashion. By using these vectors, we could prove a direct link between genetically corrected myeloid cells and the improved disease parameters observed in transplanted mice, which suggests an important role of macrophages in MSMD. Of note, macrophages, and especially tissue-resident macrophages, play a pivotal role in the progression of different diseases.47-50 The important role of macrophages is further underlined by the detection of GFP+ macrophages in the lung and by sustained lung tissue integrity after BCG infection of transplanted animals. In fact, analyzing the VCNs in transplanted animals revealed increased VCNs in lung tissue compared with bone marrow of mice that had exclusively received genetically corrected HSC, suggesting that there might be in vivo selection of macrophages after BCG infection.
The rise of modern technologies such as generation of macrophages from induced pluripotent stem cells51,52 and the recent insights into tissue-resident macrophage self-renewal and plasticity53-55 further encourage the use of modern cell types to treat the life-threatening condition of IFNγR1 deficiency in humans.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Doreen Lüttge and Theresa Buchegger (both from Hannover Medical School) for excellent technical assistance. Mycobacterium bovis Bacille Calmette-Guérin was kindly provided by Maximilliano Gutierrez (The Francis Crick Institute, London, United Kingdom).
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Cluster of Excellence REBIRTH, Exc 62/1) (N.L., R.F., and A.S.) and DFG LA 3680/2-1, Sonderforschungsbereich SFB738) (A.S.), the Else Kröner-Fresenius-Stiftung (2015_A92) (N.L.), German Centre for Infection Research (DZIF e.V. stipend) (J.S.), the Joachim Herz Stiftung (N.L.), and MHH Hannover (Young Academy program) (N.L.).
Authorship
Contribution: M.H., A.M., P.B., and N.L. designed the study, wrote the paper, and performed experiments; A.H.H.N., J.S., O.H., M.-P.K., S.B., R.M., D.B., V.H., W.B., F.-C.B., and R.G. performed experiments and analyzed data; and R.F., B.G., J.B., J.-L.C., A.S., D.J., O.H., and U.K. provided conceptual advice, discussed results, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nico Lachmann, Institute of Experimental Hematology, Hannover Medical School, Carl-Neuberg-Str 1, 30625 Hannover, Germany; e-mail: lachmann.nico@mh-hannover.de.
References
Author notes
M.H., A.M., and P.B. contributed equally to this study.


![Figure 2. Restored macrophage functionality upon Lv.SFFV.Ifnγr1 transduction of Ifnγr1−/− cells. (A) Representative histograms depicting major histocompatibility class II (MHC-II) upregulation on IFN-γ-stimulated vs unstimulated (unstim) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 3 independent experiments (two-way analysis of variance [ANOVA] using Sidak’s multiple comparisons post hoc testing. (B) Representative histograms depict CD86 upregulation on IFN-γ-stimulated vs unstimulated WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages. Bar graphs depict summarized results of 3 independent experiments (two-way ANOVA using Sidak’s multiple comparisons post hoc testing). (C) IFN-γ internalization of WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages over 24 hours (normalized to 0 hours; n = 4; two-way ANOVA using Tukey’s multiple comparisons post hoc testing). (D) Phosphorylation of STAT1 (pSTAT1) in unstimulated (–) and IFN-γ-stimulated (+) WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages compared with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control. (E) Relative messenger RNA expression of Irf1, Nos2, and Ido in IFN-γ–stimulated WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages normalized to unstimulated samples (Irf1, n = 4; Nos2 and Ido, n = 3 each). (F) Induction of ovalbumin (OVA)-specific T-cell proliferation by WT, Ifnγr1−/−, and Lv.SFFV.Ifnγr1-transduced macrophages upon stimulation with IFN-γ or IFN-γ and OVA. Bars depict the percentage of eFluor670low CD4+ T cells (n = 3; two-way ANOVA using Bonferroni’s multiple comparisons post hoc testing). (G) Bacterial burden of M avium in macrophages 0 hours and 24 hours after infection (n = 3; two-way ANOVA using Sidak’s multiple comparisons post hoc testing). (H) Bacterial burden of Bacille Calmette-Guérin colonies in macrophages 8 days after infection (normalized to uptake value 4 hours after infection; n = 3; one-way ANOVA using Dunnett’s multiple comparisons post hoc testing). *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001. For all bars, mean and standard error of the mean (SEM) are shown. ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/5/10.1182_blood-2017-10-812859/4/m_blood812859f2.jpeg?Expires=1769099350&Signature=LNpCqlazUNJERD8DZGoUKD~fBO18j284na3RQ41qv6vYUAgqcfUF4bjT6q~HqGJfFq3o~x1IVHUIQN~L9r3wZnWmmZGpCG-QcWM~zF2xZxUiy7bzC0b9jRQd0-pf2SCpqbgxdHcdDOHu3NxIfSJO4tkZbzfBcFSO45wTF8aTnnNaONtzHvm2HZof9-NMmVT3i1eCJRu1K~wpy5uEtgiyaslW8jSGAqNinEyYaBtVAFAZG-M2lOeH9MkxPJ-0-UjlzK2Ps4l9TWDQZEvIvotg1HF-kkhdQRUlkM6w97fDqrKslopMy-53Xk38wuj0dtQBUYPqz3WZSqNFo4xN8FtvkA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Engraftment of lentiviral Ifnγr1 overexpressing cells rescues Ifnγr1−/− mice from lethal pulmonary BCG infection. (A) Schematic representation of the experimental outline. Ifnγr1−/− mice were lethally irradiated and transplanted with lineage-negative Ifnγr1−/− cells transduced with Lv.SFFV.Ifnγr1 or Lv.MSP.Ifnγr1. After hematopoietic reconstitution, DsRed-labeled BCG was instilled into the lungs of the transplanted mice. Final analysis was performed 9 weeks after infection. (B) VCNs integrated into cells in the bone marrow (BM) and lungs (L) of transplanted and infected mice. Equal-size symbols represent VCNs of the same mouse in BM and L (n ≥ 3). (C) Kaplan-Meier curve depicting the survival of mice after BCG infection in 2 independent experiments (positive control [ctrl], 6 mice receiving WT cells; negative control, 13 mice receiving Lv.GFP-transduced Ifnγr1−/− cells, 7 mice receiving Lv.SFFV.Ifnγr1, and 6 mice receiving Lv.MSP.Ifnγr1. (D) IFN-γ serum levels of mice at 2, 4, 6, and 8 weeks after infection. Mean and SEM are shown for 3 independent experiments (positive control, 9 mice that received WT cells; negative control, 9 mice that received Lv.GFP-transduced Ifnγr1−/− cells and 7 nontransplanted Ifnγr1−/− mice that received Lv.SFFV.Ifnγr1 and Lv.MSP.Ifnγr1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/5/10.1182_blood-2017-10-812859/4/m_blood812859f4.jpeg?Expires=1769099350&Signature=AWk2WYBzr-TUFPEsLPPo7pyKpO3ygK~gjwZ0EGlp-Hx1WnSn0DnBHu7x0A85DJBczwGWcXTUZUIXxvUa0pNhRXjB6ghG9g9t~j~WZgQIY8AepQNr6DDnJ-ZA8rGJYKNHQ~k9C9XjvWzO5rGHfb03FCvNwLvFIrexZFap-iD7991ddpFIH1HNbWRz14rM1lBQOnBo6wfJkOigPoeG2ULYJAYDnTdLFbw3WgG4UvVBONkYD002TCtZ4l2yGI4kh3WuV5KVGQKmRRisfvd7buyV2mbX57-uJZbcavh8y37tHjjNGMmLYczFPunCJ53LJsLgte~U~B27HWOiAFmRyLYyKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Prevention of severe splenomegaly upon constitutive or myeloid-specific Ifnγr1 overexpression. (A) Representative macroscopic photos of spleens isolated from mice 9 weeks after infection. Positive control, mouse that received WT cells; negative control, nontransplanted Ifnγr1−/− mouse. (B) Spleen-to-body weight ratio depicting the normalized spleen weight of individual mice 9 weeks after infection. Mean and SEM are shown for 3 independent experiments (positive control, 10 mice that received WT cells; negative control, 13 mice that received Lv.GFP-transduced Ifnγr1−/− cells and nontransplanted Ifnγr1−/− mice; 10 mice received Lv.SFFV.Ifnγr1; 9 mice received Lv.MSP.Ifnγr1; one-way ANOVA using Tukey’s multiple comparisons post hoc testing). (C) Hematoxylin and eosin–stained histologic sections of spleens isolated 9 weeks after infection. Positive controls (mice that received WT cells), Lv.SFFV.Ifnγr1 mice, and Lv.MSP.Ifnγr1 mice showed typical spleen histology with clear separation between white pulp (areas within dotted lines [#]) and red pulp (area indicated by an asterisk), whereas this separation is absent in negative control mice (those that received Lv.GFP-transduced Ifnγr1−/− cells). Original magnification ×100. (D) BCG CFU counts of spleen homogenates 9 weeks after infection. Mean and SEM are shown for 2 independent experiments (positive control, 7 mice that received WT cells; negative control, 6 mice that received Lv.GFP-transduced Ifnγr1−/− cells; nontransplanted Ifnγr1−/− mice: Lv.SFFV.Ifnγr1, n = 4; Lv.MSP.Ifnγr1, n = 5). ***P ≤ .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/5/10.1182_blood-2017-10-812859/4/m_blood812859f5.jpeg?Expires=1769099350&Signature=rBjJDsfLMkVvcu9spJwXe-rHE3NSGPrIPyjXA9ZrYFmHpfvkDjcNh4JpxFEgdQzTfYoMvh4~XVqNrRnYevNx3hUQtYpNurFRgQhe3psnMJy2LDOHUTCZdDiveMpYO~wUXNYHuqW81scdu5M2d4vZ1B7duymiSQ-5YMfQ5wGu-xbojmWaBtjJFTNHxPejrLe7JEUu4zaMGsalKGntcAKwa46hTy3zVCc9fwHY8egT76aaQx2K8CD7bmWy5hiaPTxX6P6vLqOBMTOzu4twtIKubmLR5UOuLR5mJG4-5YjzUbhdh93g6vcvX6g5XcUEXBPe7Xxoi6m39jwAvt1KoqZSew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)