TO THE EDITOR:
RNA interference, including ectopic expression of short hairpin RNAs (shRNAs), provides a powerful approach to studying developmental pathways and genes of interest. We used this method to study Trim58, an E3 ubiquitin ligase that is highly expressed in late-stage erythroid precursors in which it facilitates degradation of the dynein molecular motor complex.1 In a widely used primary cell culture system that recapitulates terminal erythropoiesis,2 embryonic day 14.5 (E14.5) mouse (strain CD-1) fetal liver erythroblasts were expanded for 1 to 3 days in medium containing dexamethasone, stem cell factor, and erythropoietin, transduced with retroviral vectors encoding Trim58 shRNAs, and induced to undergo terminal maturation by removal of stem cell factor and dexamethasone. Compared with controls (luciferase shRNA, vector alone, or mock transduction), 4 shRNAs targeting different regions of Trim58 messenger RNA (mRNA) inhibited the transition from erythroblast to reticulocyte, a process marked by extrusion of the nucleus (enucleation).1
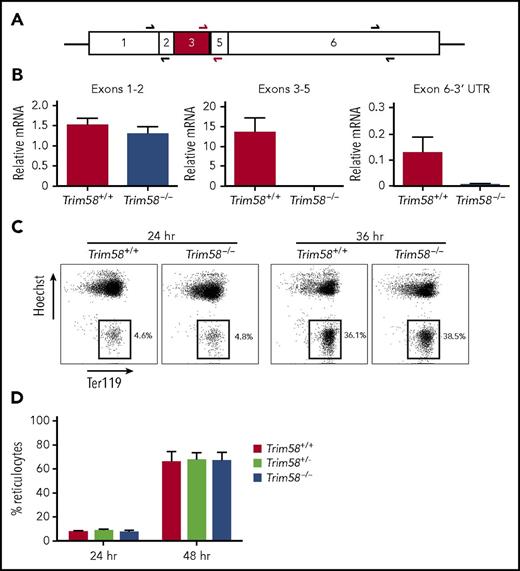
In follow-up studies described here, we disrupted the Trim58 gene in mice by removing exon 3, which encodes an essential coiled-coil domain, thereby eliminating expression of mRNA in erythroblasts (Figure 1A-B). The Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital and The Children’s Hospital of Philadelphia approved all animal protocols. Trim58−/− mice were born at Mendelian ratios and exhibited normal hemoglobin, hematocrit, and reticulocyte levels (not shown). Mutant E14.5 embryos exhibited normal external morphology, size and color, and their fetal liver erythroid precursors exhibited normal size and maturation determined by flow cytometry for Ter119, CD44, CD71, and forward scatter (not shown). Cultured E14.5 fetal liver erythroblasts from mixed strain (Sv129, FVB, and C57Bl/6) embryos exhibited normal nuclear extrusion to form reticulocytes (Figure 1C-D). In parallel studies performed under identical culture conditions, 2 independent shRNAs targeting nonoverlapping sequences of Trim58 mRNA inhibited reticulocyte formation from CD-1 erythroblasts compared with controls (Figure 2A-B), as we reported previously.1 Of note, it was not possible to rescue this phenotype by expression of shRNA-resistant Trim58 mRNA because ectopic Trim58 expression was uniformly toxic, most likely a result of nonphysiological timing of dynein degradation.
Fetal liver erythroblasts from mixed strain Trim58−/−mice mature normally. (A) Trim58 mRNA with sequences deriving from each exon indicated. Half arrows indicate positions of primer pairs used for quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of mRNA expression (panel B). Red shading indicates exon 3, which is deleted in Trim58-disrupted mice. (B) qRT-PCR analysis of E14.5 fetal liver erythroblast mRNA from wild-type (Trim58+/+) and Trim58−/− embryos using the primer pairs shown in panel A. Data are normalized to Actb mRNA and are shown as mean ± standard deviation (SD) for three biological replicate experiments. (C) E14.5 fetal liver erythroblasts isolated from mixed strain (Sv129, FVB, and C57Bl/6) Trim58+/− intercrosses were expanded in medium containing dexamethasone, stem cell factor, and erythropoietin for 2 days and then induced to undergo terminal maturation by culture in erythropoietin alone. At the indicated times after maturation induction, cells were stained with antibody against the erythroid antigen Ter119 and the membrane-permeable DNA dye Hoechst 33342 and then visualized by flow cytometry. The boxes indicate reticulocytes, which lack nuclei. (D) Reticulocytes generated after 24 and 48 hours in vitro maturation of mixed strain Trim58 gene-targeted erythroblasts, as described in panel C. Data show mean ± SD for Trim58+/+ (n = 6), Trim58+/− (n = 6), and Trim58−/− (n = 10) genotypes.
Fetal liver erythroblasts from mixed strain Trim58−/−mice mature normally. (A) Trim58 mRNA with sequences deriving from each exon indicated. Half arrows indicate positions of primer pairs used for quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of mRNA expression (panel B). Red shading indicates exon 3, which is deleted in Trim58-disrupted mice. (B) qRT-PCR analysis of E14.5 fetal liver erythroblast mRNA from wild-type (Trim58+/+) and Trim58−/− embryos using the primer pairs shown in panel A. Data are normalized to Actb mRNA and are shown as mean ± standard deviation (SD) for three biological replicate experiments. (C) E14.5 fetal liver erythroblasts isolated from mixed strain (Sv129, FVB, and C57Bl/6) Trim58+/− intercrosses were expanded in medium containing dexamethasone, stem cell factor, and erythropoietin for 2 days and then induced to undergo terminal maturation by culture in erythropoietin alone. At the indicated times after maturation induction, cells were stained with antibody against the erythroid antigen Ter119 and the membrane-permeable DNA dye Hoechst 33342 and then visualized by flow cytometry. The boxes indicate reticulocytes, which lack nuclei. (D) Reticulocytes generated after 24 and 48 hours in vitro maturation of mixed strain Trim58 gene-targeted erythroblasts, as described in panel C. Data show mean ± SD for Trim58+/+ (n = 6), Trim58+/− (n = 6), and Trim58−/− (n = 10) genotypes.
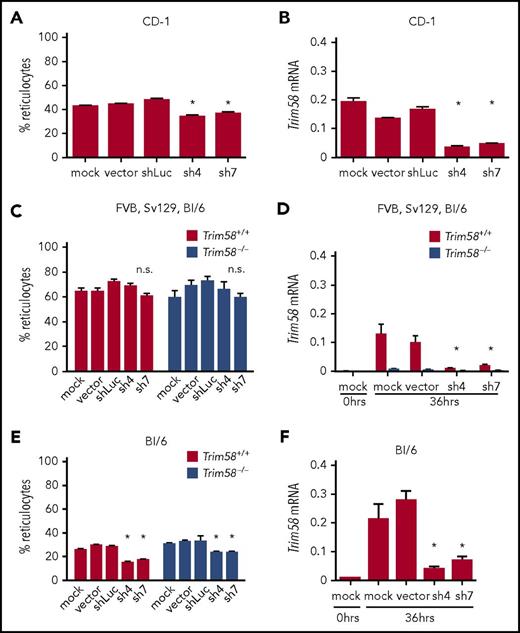
Nonspecific, genetic strain-dependent inhibition of erythroblast enucleation by Trim58 shRNAs. (A) Immature lineage (lin−) erythroid precursors from E14.5 fetal livers (CD-1 strain) were purified by immunomagnetic bead selection; expanded for 2 days in medium containing dexamethasone, stem cell factor, and erythropoietin; transduced with retroviral vectors encoding Trim58 shRNAs; then induced to undergo terminal maturation by culture in erythropoietin alone. Reticulocytes generated after 36 hours maturation were quantified as described in Figure 1C. Data show mean ± SD from three biological replicate experiments. *P < .05, 1-way analysis of variance (ANOVA). (B) Trim58 mRNA levels determined by qRT-PCR of shRNA vector-transduced CD-1 erythroblasts described in panel A after 36 hours maturation. Data are normalized to Actb mRNA and are shown as mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA. (C) Reticulocytes generated after 36 hours maturation of mixed strain Trim58+/+ and Trim58−/− erythroblasts after transduction with 2 different Trim58 shRNAs, as described in panel A. Data show mean ± SD from three biological replicate experiments, with nonsignificance (n.s.) determined by 1-way ANOVA. (D) Trim58 mRNA levels determined by qRT-PCR of mixed strain erythroblasts described in panel C after 36 hours maturation. Data are normalized as in panel B and show mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA. (E) Percent reticulocytes generated after 36 hours maturation from C57Bl/6 Trim58+/+ and Trim58−/− erythroblasts after transduction with 2 different Trim58 shRNAs. Data are presented as mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA. (F) Trim58 mRNA levels determined by qRT-PCR of C57Bl/6 erythroblasts described in panel E after 36 hours maturation. Data are normalized as in panel B and are presented as mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA.
Nonspecific, genetic strain-dependent inhibition of erythroblast enucleation by Trim58 shRNAs. (A) Immature lineage (lin−) erythroid precursors from E14.5 fetal livers (CD-1 strain) were purified by immunomagnetic bead selection; expanded for 2 days in medium containing dexamethasone, stem cell factor, and erythropoietin; transduced with retroviral vectors encoding Trim58 shRNAs; then induced to undergo terminal maturation by culture in erythropoietin alone. Reticulocytes generated after 36 hours maturation were quantified as described in Figure 1C. Data show mean ± SD from three biological replicate experiments. *P < .05, 1-way analysis of variance (ANOVA). (B) Trim58 mRNA levels determined by qRT-PCR of shRNA vector-transduced CD-1 erythroblasts described in panel A after 36 hours maturation. Data are normalized to Actb mRNA and are shown as mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA. (C) Reticulocytes generated after 36 hours maturation of mixed strain Trim58+/+ and Trim58−/− erythroblasts after transduction with 2 different Trim58 shRNAs, as described in panel A. Data show mean ± SD from three biological replicate experiments, with nonsignificance (n.s.) determined by 1-way ANOVA. (D) Trim58 mRNA levels determined by qRT-PCR of mixed strain erythroblasts described in panel C after 36 hours maturation. Data are normalized as in panel B and show mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA. (E) Percent reticulocytes generated after 36 hours maturation from C57Bl/6 Trim58+/+ and Trim58−/− erythroblasts after transduction with 2 different Trim58 shRNAs. Data are presented as mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA. (F) Trim58 mRNA levels determined by qRT-PCR of C57Bl/6 erythroblasts described in panel E after 36 hours maturation. Data are normalized as in panel B and are presented as mean ± SD for three biological replicate experiments. *P < .05 by 1-way ANOVA.
The observed differences in erythroblast enucleation could be caused by strain-specific effects of Trim58 deficiency or atypical strain-specific, off-target effects of Trim58 shRNAs. To resolve these possibilities, we transduced fetal liver erythroblasts from the progeny of intercrossed Trim58+/− mice (mixed strain or pure C57Bl/6) with Trim58 and control shRNAs. Because CD-1 mice are outbred, we could not breed the Trim58 mutant allele into this genetic background. Trim58 shRNAs did not impair reticulocyte formation from cultured mixed strain Trim58−/− or Trim58+/+ erythroblasts (Figure 2C), despite effective reduction of Trim58 mRNA in the wild-type cells (Figure 2D). In contrast, the same Trim58 shRNAs inhibited reticulocyte formation from C57Bl/6 erythroblasts by 25% to 35%, regardless of Trim58 genotype (Trim58+/+ or Trim58−/−) (Figure 2E). Both shRNAs reduced Trim58 mRNA in wild-type C57Bl/6 erythroblasts by 75% to 95% (Figure 2F). Together, these studies show that Trim58 shRNAs inhibit erythroblast enucleation through a genetic strain-specific mechanism that is independent of Trim58 expression.
shRNAs and short interfering RNAs can cause off-target effects by interacting with mRNAs through partial nucleotide sequence complementarity.3 However, this mechanism is unlikely to account for our observations, because multiple nonoverlapping Trim58 shRNAs inhibited enucleation (this study and in Thom et al1 ). More plausible explanations include shRNA-induced interferon response4 or saturation of endogenous microRNA processing pathways by high levels of viral vector–expressed shRNAs. The latter effect has been observed in hepatocytes partly via inhibition of exportin-5, which mediates nuclear export of shRNAs and microRNAs.5 Consistent with this mechanism, exportin-5 declines progressively during mouse erythropoiesis,6 which could render late-stage erythroblasts more susceptible to ectopic shRNA-induced depletion of endogenous microRNAs that are essential for terminal maturation.7,8 In current studies, erythroblasts were transduced by a green fluorescent protein–marked shRNA retroviral vector at low multiplicity of infection (0.3 to 0.5) and then selected under puromycin to achieve ∼1 vector copy per cell. However, different shRNAs encoded by the same vector can vary considerably in their expression levels and consequent nonspecific, dosage-dependent effects on cellular phenotypes.5 Although further studies are required to define the mechanism by which Trim58 shRNAs inhibit erythroblast enucleation nonspecifically in certain mouse strains, our findings indicate that the effects of any shRNA on late-stage erythroid maturation must be interpreted cautiously and validated through independent approaches. These could include alternate vector systems that allow for better titration and monitoring of shRNA expression,9 phenotypic rescue by shRNA-resistant forms of the targeted mRNA (not possible for this study), or genome editing.10
Acknowledgments
The authors thank Mark Kahn and Gerd Blobel for helpful comments on the manuscript.
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants R01 DK092318 (M.J.W.), R01 DK61692 (M.J.W.), and 5F30DK102291-02 (E.A.T.). Murine stem cell virus plasmid (puromycin-internal ribosomal entry site-green fluorescent protein) (Addgene plasmid #18751) was a gift from Scott Lowe.
Authorship
Contribution: All authors designed experiments; E.A.T., C.S.T., Y.Y., and V.P. performed experiments; and E.A.T and M.J.W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mitchell J. Weiss, Department of Hematology, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, MS#355, Memphis, TN 38105-3678; e-mail: mitch.weiss@stjude.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal