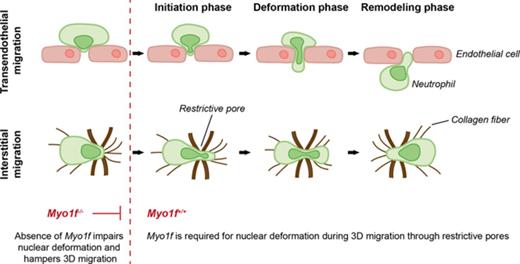
Key Points
Myo1f is critical for migration in 3D environments.
Myo1f regulates the dynamic deformation of the nucleus during migration through physical barriers.
Abstract
Neutrophil extravasation and interstitial migration are important steps during the recruitment of neutrophils to sites of inflammation. In the present study, we addressed the functional importance of the unconventional class I myosin 1f (Myo1f) for neutrophil trafficking during acute inflammation. In contrast to leukocyte rolling and adhesion, the genetic absence of Myo1f severely compromised neutrophil extravasation into the inflamed mouse cremaster tissue when compared with Myo1f+/+ mice as studied by intravital microscopy. Similar results were obtained in experimental models of acute peritonitis and acute lung injury. In contrast to 2-dimensional migration, which occurred independently of Myo1f, Myo1f was indispensable for neutrophil migration in 3-dimensional (3D) environments, that is, transmigration and migration in collagen networks as it regulated squeezing and dynamic deformation of the neutrophil nucleus during migration through physical barriers. Thus, we provide evidence for an important role of Myo1f in neutrophil trafficking during inflammation by specifically regulating neutrophil extravasation and migration in 3D environments.
Introduction
Polymorphonuclear neutrophils as important players of the innate immune system represent the first leukocytes recruited to sites of injury during an acute inflammatory response following a well-defined multistep cascade of adhesion and activation events.1,2 After initial capturing and rolling of neutrophils on the inflamed endothelium, the subsequent steps include slow rolling, induction of adhesion, adhesion strengthening, spreading, intraluminal crawling, transendothelial migration, and abluminal crawling.1,3 During extravasation, neutrophils traverse the endothelial barrier via the paracellular or to a lesser extent via the transcellular route before passing the basement membrane and pericytes to enter the interstitial space.4-7 Subsequent migration within 3-dimensional (3D) environments occurs in the context of, for example, a fibrillary network in the interstitial space or a cell-packed environment in case of organ parenchymas.8
In general, migration requires actin polymerization to push the leading membrane forward and actomyosin contraction to detach the trailing edge.9-11 For successful cell migration in 3D environments, that is, transmigration and interstitial migration, the nucleus must undergo defined changes in position and shape, which is dependent on cytoskeletal dynamics at the leading and trailing edge to push the nucleus through restrictive barriers.12-14 During migration in 2-dimensional (2D) environments, including intraluminal crawling, the deformation of the nucleus is not critical due to the absence of restriction sites.9 The deformability of the nucleus during migration in 3D environments depends on dynamic interaction between the actin cytoskeleton and the nucleus, which relies on the mechanical linkage between the actin filaments and the nuclear membrane.15-17
The unconventional class I myosins consist of a motor domain with an actin-binding site, followed by a light-chain–binding IQ motif and a basic tail homology 1 (TH1) domain that is essential for interactions with membranes.18 Thus, class I myosins may putatively link the actin cytoskeleton to the nuclear membrane enabling the transmission of force from the actin cytoskeleton to the nucleus.15,17 Previous studies identified the unconventional class I myosin 1f (Myo1f) to be important for different membrane-associated functions including endocytosis, cell signaling, and cell motility.19-21 Myo1f was found to be mainly expressed in neutrophils, and the genetic absence of Myo1f caused defects in extravasation and host defense that were attributed to increased neutrophil adhesion via β2 integrins under static conditions in vitro.22 In the present study, we addressed the functional importance of Myo1f for neutrophil trafficking during acute inflammation in detail. In line with previous studies,22 we observed increased adhesion under static conditions in vitro, however, induction of adhesion, spreading, and migration under physiological shear stress conditions was not significantly altered in the genetic absence of Myo1f. These results were confirmed using intravital microscopy where no differences of neutrophil rolling and adhesion were observed in tumor necrosis factor α (TNFα)-stimulated cremaster muscle venules of Myo1f−/− and Myo1f+/+ mice. Strikingly, the genetic absence of Myo1f severely compromised neutrophil extravasation into the inflamed mouse cremaster tissue. Similarly, neutrophil extravasation was dramatically reduced in models of acute peritonitis and lung injury. The genetic absence of Myo1f significantly impaired neutrophil transmigration and migration in collagen gels. Detailed live cell imaging revealed that Myo1f was important for the dynamic deformation of the neutrophil nucleus during migration through physical barriers. Thus, Myo1f is specifically required for migration in 3D environments during neutrophil trafficking in innate immunity.
Methods
Animals
Myo1f−/− and Myo1f+/+ mice were maintained on a C57BL/6 background.22 C57BL/6 wild type (Myo1f+/+) mice were obtained from Charles River Laboratories. All animal experiments were conducted in accordance with the German federal animal protection laws and approved by the Bavarian Government (Regierung von Oberbayern, Munich, Germany).
Live cell imaging of in vitro transmigration under flow
Transmigration of neutrophils was analyzed in μ-Slide membrane ibiPore flow chambers (Ibidi) consisting of a flow chamber, a 300-nm-thick membrane with 5-µm pores, and a subjacent collagen gel (1.5 mg/mL) containing 10 µM N-formylmethionyl-leucyl-phenylalanine (fMLP). For F-actin staining, neutrophils were labeled with silicon rhodamin (SiR)–actin23 (100 nM; Spirochrome) 24 hours prior to the experiment as described24 and perfused through the recombinant murine (rm)ICAM-1 (12.5 µg/mL) and rmP-selectin (10 µg/mL) coated flow chamber with 1 dyne/cm2 shear stress for 60 minutes. Time-lapse video microscopy with an average interval of 30 seconds was performed using an upright spinning-disk confocal microscope (Examiner; Zeiss) equipped with a confocal scanner unit CSU-X1 (Yokogawa Electric Corporation, Japan), an EMCCD camera (Evolve; Photometrics); and a 20×/1.0 NA water immersion objective (Plan Apochromat; Zeiss). 3D images (70 z-stacks with a step size of 0.55 µm) were acquired, and transmigration was analyzed by generating orthogonal projection of z-stacks over time using Slidebook 6.0.8 software (3i).
3D chemotaxis
Analysis of 3D chemotaxis was performed in Ibidi µ-Slides Chemotaxis 3D chambers according to the manufacturer’s protocol. Briefly, isolated murine neutrophils were seeded into gels with different concentrations of rat tail collagen type I (1.5, 3.0 mg/mL) and exposed to a gradient of CXCL1 (100 ng/mL). Time-lapse video microscopy was performed every 14 seconds at 37°C for 30 minutes using an Axiovert 200M microscope. In a second set of experiments, the neutrophil nuclei were stained with the nuclear dye Hoechst 33342 (5 µM) and 3D chemotaxis was performed in a 1.5 mg/mL collagen gel with a CXCL1 (100 ng/mL) chemoattractant gradient. The shape of the nucleus during 3D migration was analyzed using an upright spinning-disk confocal microscope (Examiner; Zeiss), as described in the previous section. The mean nuclear shape change factor per field of view per time point was analyzed using ImageJ’s plugin Moment Calculator provided by Francois Richard (University of Ottawa, Ottawa, ON, Canada) and normalized to the overall smallest value to calculate the percentage change in nuclear shape within the following 10 minutes of observation. The shape of the nucleus and the subcellular localization of Myo1f and Actin were analyzed during 3D chemotaxis in a 1.5 mg/mL collagen gel with fMLP (100 nM) as chemoattractant using dHL-60-EGFP-Myo1f cells and live cell spinning-disk confocal microscopy as mentioned in the previous section. Briefly, dHL-60-EGFP-Myo1f cells were stained with the nuclear dye Hoechst 33342 (5 µM) for 5 minutes at 37°C and for F-actin with SiR-actin (100 nM; Spirochrome) 24 hours prior to the experiment. Images were acquired using Slidebook 6.0.8 software and by using 3 lasers with an excitation wavelength of 405 nm, 488 nm, and 640 nm.
Statistical analysis
All data were analyzed and plotted using GraphPad Prism 6 software (GraphPad Software Inc) and are shown as means ± standard error of the mean (SEM). Statistical significance for pairwise comparison of experimental groups was determined using an unpaired Student t test. For multiple comparisons, 2-way analysis of variance (ANOVA) with Sidak multiple comparisons test (comparison of all experimental groups against each other) was used. P <.05 was considered as statistically significant.
Supplemental methods
Further in vitro and in vivo experiments including intravital microscopy,25,26 lipopolysaccharide (LPS)-induced lung injury,27-29 neutrophil adhesion, spreading and migration assays24-26,30,31 as well as transmigration and generation of the HL-60-EGFP-Myo1f cell line by lentiviral transduction31,32 are described in detail in supplemental Methods (available on the Blood Web site).
Results
Myo1f was dispensable for leukocyte rolling and adhesion under physiological flow conditions but fundamentally important for neutrophil extravasation
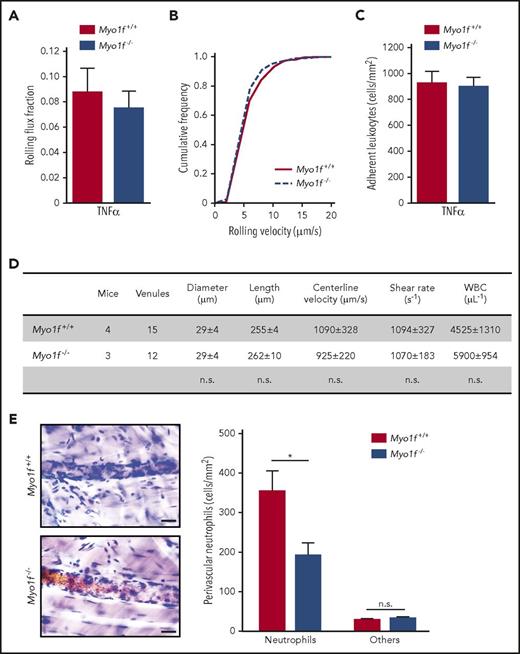
To study the biological relevance of Myo1f for neutrophil trafficking, intravital microscopy was performed using Myo1f+/+ and Myo1f−/− mice. In TNFα-stimulated cremaster muscle venules, leukocyte rolling flux fraction, rolling velocity as well as the number of adherent leukocytes were similar between the 2 mouse strains (Figure 1A-C). This was also true upon systemic CXCL1 injection, indicating that the genetic absence of Myo1f did not alter β2 integrin–mediated neutrophil adhesion to inflamed endothelial cells in vivo33 (supplemental Figure 1A-B). The differential white blood cell counts (data not shown) as well as hemodynamic and microvascular parameters were similar between both groups (Figure 1D; supplemental Figure 1C). In line with these findings, induction of neutrophil adhesion in vitro was unaffected in Myo1f−/− neutrophils compared with Myo1f+/+ neutrophils under physiological flow conditions (1 dyne/cm2) when exposed to rmP-selectin, rmICAM-1, and CXCL1 (supplemental Figure 2A). This finding was in sharp contrast to the situation under static conditions in vitro, where Myo1f-deficient neutrophils showed increased adhesion to the β2 integrin ligand ICAM-1 as demonstrated earlier by Kim et al22 (supplemental Figure 2B). Thus, in accordance with the literature, Myo1f affected adhesion under static conditions in vitro22 but not under physiological flow conditions in vivo and in vitro.
Myo1f was dispensable for leukocyte rolling and adhesion but essential for neutrophil extravasation in inflamed mouse cremaster muscle venules. (A-C) Intravital microscopy of postcapillary venules in mouse cremaster muscle 2.5 hours after intrascrotal injection of TNFα (500 ng per animal). (A) Rolling flux fraction, (B) rolling velocity, and (C) number of adherent leukocytes (n = 15 venules of 4 Myo1f+/+ mice and n = 12 venules of 3 Myo1f−/− mice). (D) Rheological parameters of cremaster muscle venules after TNFα injection. (E) Representative images (left panel) of postcapillary venules of cremaster muscle whole mounts stained with Giemsa’s azur eosin methylene blue. Scale bar, 20 µm. Quantification of perivascular neutrophils and other leukocyte subtypes (others, right panel) (n = 18 vessels of 3 Myo1f+/+ mice and n = 17 vessels of 3 Myo1f−/− mice). Mean ± SEM; *P < .05 (2-way ANOVA, Sidak multiple comparison test). n.s., not significant; WBC, white blood cell.
Myo1f was dispensable for leukocyte rolling and adhesion but essential for neutrophil extravasation in inflamed mouse cremaster muscle venules. (A-C) Intravital microscopy of postcapillary venules in mouse cremaster muscle 2.5 hours after intrascrotal injection of TNFα (500 ng per animal). (A) Rolling flux fraction, (B) rolling velocity, and (C) number of adherent leukocytes (n = 15 venules of 4 Myo1f+/+ mice and n = 12 venules of 3 Myo1f−/− mice). (D) Rheological parameters of cremaster muscle venules after TNFα injection. (E) Representative images (left panel) of postcapillary venules of cremaster muscle whole mounts stained with Giemsa’s azur eosin methylene blue. Scale bar, 20 µm. Quantification of perivascular neutrophils and other leukocyte subtypes (others, right panel) (n = 18 vessels of 3 Myo1f+/+ mice and n = 17 vessels of 3 Myo1f−/− mice). Mean ± SEM; *P < .05 (2-way ANOVA, Sidak multiple comparison test). n.s., not significant; WBC, white blood cell.
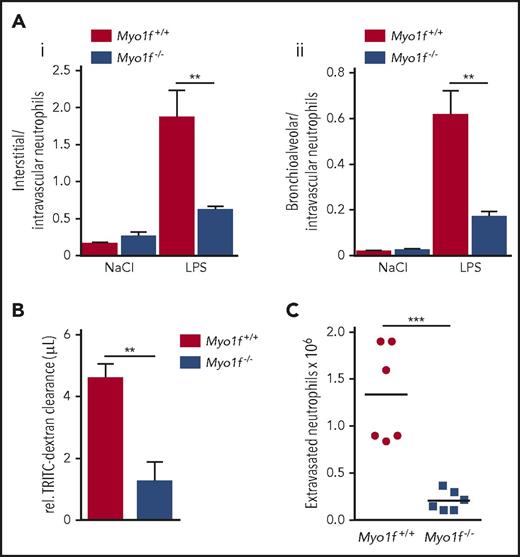
Next, we examined the ability of Myo1f-deficient neutrophils to emigrate into inflamed mouse cremaster tissue upon intrascrotal injection of TNFα. Histological analysis revealed that the number of perivascular neutrophils (but not other leukocytes) was decreased in Myo1f−/− mice compared with Myo1f+/+ animals, demonstrating that Myo1f was critical for neutrophil extravasation into inflamed cremaster tissue (Figure 1E). To verify the importance of Myo1f for neutrophil extravasation in inflammation, we induced an acute lung injury in mice by exposure to aerosolized LPS or NaCl (negative control) for 30 minutes and subsequently quantified intravascular, interstitial, and alveolar myeloid cells after 4 hours. Similar to the in vitro data under static conditions, the primary data revealed increased intravascular neutrophil accumulation in Myo1f−/− compared with Myo1f+/+ mice upon LPS inhalation under low shear stress conditions in the lung vasculature, whereas neutrophil numbers in the interstitial space and the bronchoalveolar lavage were reduced in the genetic absence of Myo1f (supplemental Figure 3). To analyze the extravasation efficiency of neutrophils, we calculated the ratio of interstitial/intravascular neutrophils as well as bronchioalveolar/intravascular neutrophils. In Myo1f+/+ mice, extravasation of neutrophils from the lung vasculature into the interstitium (Figure 2Ai) and the bronchoalveolar space (Figure 2Aii) was dramatically increased in LPS-treated mice compared with NaCl-treated controls as expected. In contrast, LPS-induced extravasation into the interstitium as well as into the bronchoalveolar space was significantly impaired in Myo1f−/− mice compared with Myo1f+/+ mice. Accordingly, LPS-induced lung injury analyzed by measuring vascular permeability was significantly decreased in the genetic absence of Myo1f (Figure 2B). Furthermore, the ability of Myo1f−/− neutrophils to extravasate in acute peritonitis induced by intraperitoneal application of CXCL1 was almost completely abolished compared with Myo1f+/+ mice (Figure 2C). Thus, the genetic absence of Myo1f impaired the capacity of neutrophils to extravasate into the inflamed tissue, including the cremaster muscle, the lungs, and the peritoneal cavity.
Myo1f-deficient neutrophils failed to extravasate in an acute lung injury model and during acute peritonitis. (A) Ratios of Myo1f+/+ and Myo1f−/− neutrophils extravasated into the interstitium (i) or the bronchoalveolar space (ii) to neutrophils remaining in the lung vasculature, 4 hours upon exposure to aerosolized NaCl or LPS for 30 minutes. n = 4; mean ± SEM; **P < .01 (2-way ANOVA, Tukey multiple comparison test). (B) Lung damage was assessed by quantification of tetramethylrhodamine isothiocyanate (TRITC)-Dextran clearance. n = 4; mean ± SEM; **P < .01 (unpaired Student t test). (C) Total number of extravasated neutrophils in the peritoneal lavage 4 hours after intraperitoneal injection of CXCL1 (300 ng per animal). n = 6; mean ± SEM; ***P < .001 (unpaired Student t test).
Myo1f-deficient neutrophils failed to extravasate in an acute lung injury model and during acute peritonitis. (A) Ratios of Myo1f+/+ and Myo1f−/− neutrophils extravasated into the interstitium (i) or the bronchoalveolar space (ii) to neutrophils remaining in the lung vasculature, 4 hours upon exposure to aerosolized NaCl or LPS for 30 minutes. n = 4; mean ± SEM; **P < .01 (2-way ANOVA, Tukey multiple comparison test). (B) Lung damage was assessed by quantification of tetramethylrhodamine isothiocyanate (TRITC)-Dextran clearance. n = 4; mean ± SEM; **P < .01 (unpaired Student t test). (C) Total number of extravasated neutrophils in the peritoneal lavage 4 hours after intraperitoneal injection of CXCL1 (300 ng per animal). n = 6; mean ± SEM; ***P < .001 (unpaired Student t test).
Loss of Myo1f did not impair neutrophil spreading and migration in 2D environments
To identify the underlying cellular defect causing diminished extravasation, neutrophil spreading, polarization, and migration were analyzed in vitro in flow chambers using isolated murine Myo1f+/+ and Myo1f−/− neutrophils. Under physiological shear stress conditions (1 dyne/cm2), spreading and polarization of Myo1f−/− neutrophils on immobilized fibrinogen or rmICAM-1 were normal compared with Myo1f+/+ neutrophils (supplemental Figure 4A). Similarly, analysis of mechanotactic migration on immobilized fibrinogen in the presence of fMLP or immobilized rmICAM-1 and CXCL1 revealed no difference regarding the Euclidean distance and migration velocity of Myo1f−/− neutrophils compared with Myo1f+/+ neutrophils (supplemental Figure 5A). Next, we analyzed the capacity of Myo1f−/− neutrophils to polarize, spread, and migrate in the presence of a chemotactic gradient (fMLP or CXCL1) using Zigmond chambers. Similar to flow conditions, spreading and polarization was not altered in the genetic absence of Myo1f upon exposure to immobilized fibrinogen or rmICAM-1 (supplemental Figure 4B). Furthermore, chemotactic migration of neutrophils, that is, the Euclidean distance and migration velocities in response to fMLP or CXCL1 was normal in the genetic absence of Myo1f (supplemental Figure 5B). In conclusion, Myo1f was dispensable for spreading, polarization, and mechanotactic migration under flow conditions as well as chemotactic migration in 2D environments suggesting that the increased adhesiveness observed under static was not responsible for the extravasation defect.
Myo1f was crucial for neutrophil transmigration
Next, we studied the process of transmigration under physiological flow conditions (1 dyne/cm2) in vitro using μ-Slide membrane ibiPore flow chambers and spinning-disk confocal microscopy. This device consisted of a 300-nm-thick membrane with a transwell filter coated with rmICAM-1 and rmP-selectin, separating the flow chamber from the collagen gel, which contained 10 µM fMLP as chemoattractant. Myo1f+/+ and Myo1f−/− neutrophils were perfused through the flow chamber with 1 dyne/cm2 shear stress and transmigration was analyzed in real-time (4-dimensional). Figure 3A demonstrates successful transmigration of Myo1f+/+ neutrophils into the lower collagen compartment after 60 minutes of stimulation. In contrast, Myo1f-deficient neutrophils showed defective transmigration and the majority of neutrophils were found within the flow chamber or stuck within the pores. For detailed analysis, neutrophils were quantified according to their localization, that is, neutrophils located within the flow chamber (flow chamber), stuck within the pores (pores), or fully transmigrated into the collagen gel (collagen gel). Here, 60% ± 12% Myo1f+/+ neutrophils transmigrated into the collagen gel. Only 11% ± 9% cells remained within the flow chamber, and 29% ± 8% neutrophils were located within the pore after stimulation for 60 minutes. This was in sharp contrast to Myo1f−/− neutrophils, where only 13% ± 7% transmigrated into the collagen gel and the majority of cells were located within the pore (63% ± 18%) or remained in the flow chamber (25% ± 15%; Figure 3B). In a second set of experiments, we performed transwell assays where filters with a relatively large pore size of 8 µm were coated with a monolayer of b.End3 cells. Similar to the findings under physiological flow conditions, stimulation of Myo1f+/+ neutrophils with CXCL1 induced robust and highly significant transmigration compared with unstimulated Myo1f+/+ neutrophils. Whereas, this response was almost completely absent in Myo1f−/− neutrophils (Figure 3C). Using uncoated filters as control, CXCL1 stimulation induced sustained transmigration of Myo1f+/+ neutrophils whereas Myo1f−/− neutrophils were almost completely unable to transmigrate, indicating that Myo1f was critical for neutrophil transmigration through physical barriers (Figure 3D).
Myo1f was required for neutrophil transmigration. (A-B) Myo1f+/+ and Myo1f−/− neutrophils were stained with SiR-actin (100 nM) and perfused through rmICAM-1 (12.5 µg/mL) and rmP-selectin (10 µg/mL) coated μ-Slide Membrane ibiPore flow chambers with 1 dyne/cm2 shear stress. Time-lapse video microscopy was performed using spinning-disk confocal microscopy. (A) Orthogonal view of representative pseudo-colored time-lapse images demonstrating the localization of neutrophils during the process of transmigration into a fMLP-containing collagen chamber after stimulation for 60 minutes. Scale bar, 10 µm. Color scales, heat map. Triangle indicates orientation of fMLP gradient. Arrow indicates direction of flow. (B) Quantitative analysis of transmigrating Myo1f+/+ and Myo1f−/− neutrophils respective to their position in percentage of all neutrophils analyzed (100%). n = 4 independent experiments (with a total of 240 Myo1f+/+ neutrophils and 220 Myo1f−/− neutrophils analyzed). Mean ± SEM; ***P < .001; ****P < .0001 (2-way ANOVA, Sidak multiple comparison test). (C-D) Transwell assay without (−) or with CXCL1 either coated with (C) an endothelial monolayer, or (D) uncoated as control. n = 4; mean ± SEM; ****P < .0001.
Myo1f was required for neutrophil transmigration. (A-B) Myo1f+/+ and Myo1f−/− neutrophils were stained with SiR-actin (100 nM) and perfused through rmICAM-1 (12.5 µg/mL) and rmP-selectin (10 µg/mL) coated μ-Slide Membrane ibiPore flow chambers with 1 dyne/cm2 shear stress. Time-lapse video microscopy was performed using spinning-disk confocal microscopy. (A) Orthogonal view of representative pseudo-colored time-lapse images demonstrating the localization of neutrophils during the process of transmigration into a fMLP-containing collagen chamber after stimulation for 60 minutes. Scale bar, 10 µm. Color scales, heat map. Triangle indicates orientation of fMLP gradient. Arrow indicates direction of flow. (B) Quantitative analysis of transmigrating Myo1f+/+ and Myo1f−/− neutrophils respective to their position in percentage of all neutrophils analyzed (100%). n = 4 independent experiments (with a total of 240 Myo1f+/+ neutrophils and 220 Myo1f−/− neutrophils analyzed). Mean ± SEM; ***P < .001; ****P < .0001 (2-way ANOVA, Sidak multiple comparison test). (C-D) Transwell assay without (−) or with CXCL1 either coated with (C) an endothelial monolayer, or (D) uncoated as control. n = 4; mean ± SEM; ****P < .0001.
Myo1f was required for migration in 3D environments by squeezing the nucleus through narrow spaces
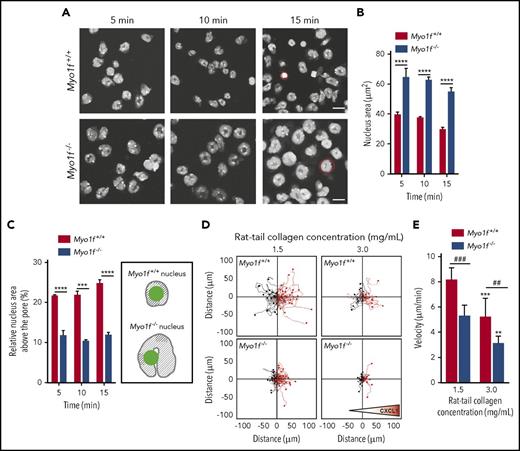
To analyze the putative role of Myo1f for squeezing the neutrophil through narrow spaces, we performed transwell assays and analyzed the morphology of the nucleus during transmigration through rmICAM-1 and rmP-selectin coated transwell filters upon stimulation with fMLP. Confocal microscopy revealed that, upon fMLP stimulation for 5 minutes, the cross-sectional area of the nuclei of Myo1f+/+ neutrophils was 39.3 ± 3 µm2. Strikingly, the area of the nuclei of Myo1f−/− neutrophils was 64.3 ± 6 µm2 and thus significantly larger than the nuclei of Myo1f+/+ neutrophils (Figure 4A-B), whereas cytospins of freshly isolated Myo1f+/+ and Myo1f−/− neutrophils revealed a similar area of the nuclei in both cell populations (supplemental Figure 6). Similar effects were observed at later time points (nucleus area of Myo1f+/+ neutrophils: 37.2 ± 1 µm2 after 10 minutes and 30.5 ± 2 µm2 after 15 minutes, nucleus area of Myo1f−/− neutrophils: 62.3 ± 3 µm2 after 10 minutes and 55.5 ± 4 µm2 after 15 minutes). In addition, the relative nucleus area located above the pores in percentage of the whole nucleus area (100%) was significantly larger in Myo1f+/+ neutrophils compared with Myo1f−/− neutrophils (Figure 4C), suggesting that Myo1f is critical for squeezing the nucleus through restrictive sites.
Myo1f was crucial for squeezing of the neutrophil nucleus through narrow pores during migration in 3D environments. (A-C) Nucleus morphology was analyzed in transwell assays with rmICAM-1 (12.5 µg/mL) and rmP-selectin (10 µg/mL) coated filters upon fMLP stimulation (10 µM). Isolated murine Myo1f+/+ and Myo1f−/− neutrophils were labeled with the nuclear dye Hoechst 33342 (5 µM) and imaged at indicated time points. (A) Representative microscopic images of nuclear morphology on the coated filters at indicated time points. Red circles indicate a representative nucleus of a Myo1f+/+ and a Myo1f−/− neutrophil. Scale bar, 10 µm. (B) Quantification of the total area of the nuclei as well as (C) quantification and schematic illustration of the relative area of the nuclei above the pore upon stimulation of Myo1f+/+ and Myo1f−/− neutrophils with fMLP for 5, 10, and 15 minutes. In the schematic representation, the nuclei are indicated in gray and the pore in green. The area of the Myo1f−/− nuclei is larger than the area of the Myo1f+/+ nuclei due to defective nucleus deformation in Myo1f−/− neutrophils. Thus, the relative nucleus area located above/within the pore (green) is larger in Myo1f+/+ neutrophils as a smaller area of the nucleus is not located above/within the pore (shaded area). n = 3 independent experiments with ∼76 Myo1f+/+ and ∼78 Myo1f−/− neutrophils at each time point. Mean ± SEM; **P < .01; ***P < .001; ****P < .0001 (2-way ANOVA, Sidak multiple comparison test). (D-E) 3D chemotactic migration of Myo1f+/+ and Myo1f−/− neutrophils toward a CXCL1 (100 ng/mL) gradient using 3D chemotaxis chambers. (D) Single-cell migration tracks in 3D collagen gels with different collagen concentrations. Triangle indicates orientation of the gradient. (E) Quantitative analysis of mean migration velocity. n = 4; mean ± SEM; ##P < .01; ###P < .001; **P < .01; ***P < .001 vs rat tail collagen concentration 1.5 mg/mL (2-way ANOVA, Sidak multiple comparison test).
Myo1f was crucial for squeezing of the neutrophil nucleus through narrow pores during migration in 3D environments. (A-C) Nucleus morphology was analyzed in transwell assays with rmICAM-1 (12.5 µg/mL) and rmP-selectin (10 µg/mL) coated filters upon fMLP stimulation (10 µM). Isolated murine Myo1f+/+ and Myo1f−/− neutrophils were labeled with the nuclear dye Hoechst 33342 (5 µM) and imaged at indicated time points. (A) Representative microscopic images of nuclear morphology on the coated filters at indicated time points. Red circles indicate a representative nucleus of a Myo1f+/+ and a Myo1f−/− neutrophil. Scale bar, 10 µm. (B) Quantification of the total area of the nuclei as well as (C) quantification and schematic illustration of the relative area of the nuclei above the pore upon stimulation of Myo1f+/+ and Myo1f−/− neutrophils with fMLP for 5, 10, and 15 minutes. In the schematic representation, the nuclei are indicated in gray and the pore in green. The area of the Myo1f−/− nuclei is larger than the area of the Myo1f+/+ nuclei due to defective nucleus deformation in Myo1f−/− neutrophils. Thus, the relative nucleus area located above/within the pore (green) is larger in Myo1f+/+ neutrophils as a smaller area of the nucleus is not located above/within the pore (shaded area). n = 3 independent experiments with ∼76 Myo1f+/+ and ∼78 Myo1f−/− neutrophils at each time point. Mean ± SEM; **P < .01; ***P < .001; ****P < .0001 (2-way ANOVA, Sidak multiple comparison test). (D-E) 3D chemotactic migration of Myo1f+/+ and Myo1f−/− neutrophils toward a CXCL1 (100 ng/mL) gradient using 3D chemotaxis chambers. (D) Single-cell migration tracks in 3D collagen gels with different collagen concentrations. Triangle indicates orientation of the gradient. (E) Quantitative analysis of mean migration velocity. n = 4; mean ± SEM; ##P < .01; ###P < .001; **P < .01; ***P < .001 vs rat tail collagen concentration 1.5 mg/mL (2-way ANOVA, Sidak multiple comparison test).
In addition to transmigration, 3D migration in fibrillary networks requires rapid change of the cell shape as well as deformation of the nucleus for effective migration.34 To study the functional impact of Myo1f for 3D migration, we performed 3D chemotaxis experiments using different concentrations of collagen and analyzed the migration velocity of Myo1f+/+ and Myo1f−/− neutrophils toward a CXCL1 gradient in 3D collagen gels (Figure 4D-E). As expected,9,34 we found the accumulated migration distance, that is, the migration velocity of Myo1f+/+ neutrophils to be reduced in a high-density collagen gel (3 mg/mL) compared with a low-density collagen gel (1.5 mg/mL). In a 1.5 mg/mL collagen gel, Myo1f−/− neutrophils showed a significantly reduced migration velocity compared with Myo1f+/+ neutrophils. In high-density collagen (3 mg/mL), the migration capacity of Myo1f−/− neutrophils was even further diminished compared with Myo1f+/+ neutrophils (Figure 4D-E). Rose diagrams indicated that sensing of the CXCL1 gradient was still intact in Myo1f-deficient neutrophils (supplemental Figure 7). Thus, our data demonstrated that in contrast to 2D migration, which was unaffected in the genetic absence of Myo1f, Myo1f was indispensable for 3D migration.
Myo1f regulated the nuclear shape change during migration in 3D environments
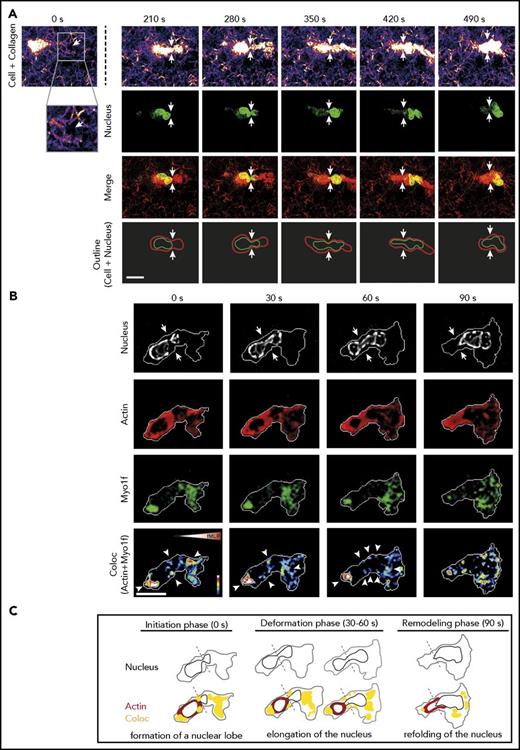
To visualize the restrictive barriers in the collagen meshwork that neutrophils pass while migrating within this confined 3D environment, live cell imaging was performed using confocal reflection/fluorescence microscopy (supplemental Video 1). Figure 5A shows the still images of a representative primary human neutrophil migrating through a collagen meshwork. As expected, migration of the human neutrophil through a restrictive barrier (white arrows) was initiated by the formation of a small nuclear lobe preparing the deformation of the nucleus35 (280 seconds). This step was followed by further deformation and elongation of the nucleus (350-420 seconds). Having completed squeezing of the nucleus through the narrow pore, the nucleus was refolded into a roundish, multilobular shape (490 seconds).34 To analyze the interplay between Myo1f and the nucleus during 3D migration in collagen gels, we generated HL-60 cells stably expressing EGFP-Myo1f (supplemental Figure 8). Similar to human neutrophils, the nucleus of dHL-60 EGFP-Myo1f cells formed a nuclear lobe (0 seconds), elongated and deformed (30-60 seconds) to squeeze the cell through constriction sites while the cell migrated through the collagen gel (Figure 5B). Actin formed a ring-like structure around the nucleus during the initiation and deformation phases (0-60 seconds). This ring-like structure was not observed during migration in 2D environments (data not shown). However, during migration in 3D environments, Myo1f was mainly accumulated at the rear and the front of the cell. After passing the narrow pore, an open actin ring remained at the rear of the transmigrated cell (remodeling phase, 90 seconds), whereas Myo1f was distributed throughout the cell. Importantly, at the constriction sites, as well as at the rear and the front of the cell, we observed colocalization of Myo1f and Actin (coloc, white arrowheads) during the initiation and deformation phases (0-60 seconds) pointing toward the important role of Myo1f for bringing the nucleus in shape for migration through narrow pores. Figure 5C shows the schematics of nuclear deformation of the dHL-60 EGFP-Myo1f cell in Figure 5B during migration in 3D environments including the initiation phase (0 seconds), the deformation phase (30-60 seconds), and the remodeling phase (90 seconds). Similar results were obtained using isolated human neutrophils (supplemental Figure 9).
Live cell imaging of nuclear deformation and dynamics of Actin and Myo1f localization during migration in 3D environments. Analysis of 3D migration in a 1.5 mg/mL collagen gel toward an fMLP (100 nM) gradient was performed with dHL-60-EGFP-Myo1f cells or isolated human neutrophils. (A) Live cell imaging of the human neutrophil nucleus (labeled with Hoechst 5 µM, green) and the collagen meshwork (fire and red) using confocal reflection/fluorescence microscopy. Serial images of a movie of a representative neutrophil migrating through a meshwork of collagen fibers at indicated time points. Confocal reflection images demonstrate the architecture of the collagen gel as well as the neutrophil body (fire and red). Fluorescence images depict the shape of the nucleus during migration within a collagen gel (green). Merge (yellow) shows the reflection image (red) and the nucleus (green). Schematic outline of the shape of the cell (red) and the nucleus (green) while the neutrophil squeezes through a narrow pore. Arrows indicate the pore (white). Neutrophil localization is normalized to the position of the pore. Scale bar, 10 µm. The cell is representative for a total of 12 cells from 3 independent experiments. (B) Live cell imaging of a migrating dHL-60-EGFP-Myo1f cell using spinning-disk confocal microscopy. Pseudo-colored images demonstrate the morphology of the nucleus (gray) of dHL-60 EGFP-Myo1f cells, the subcellular localization of Actin (red) and Myo1f (green), as well as the colocalization of Actin and Myo1f (arrowheads). Arrows indicate constriction site. Scale bar, 10 µm. Color scales, heat map. Triangle indicates orientation of fMLP gradient. The cell is representative for a total of 9 cells from 3 independent experiments. (C) Schematic outline of the nuclear morphology (black), localization of Actin (red) and Myo1f (green), and colocalization of Actin and Myo1f (yellow) during 3D migration at indicated time points. The border of the cell body is depicted in gray.
Live cell imaging of nuclear deformation and dynamics of Actin and Myo1f localization during migration in 3D environments. Analysis of 3D migration in a 1.5 mg/mL collagen gel toward an fMLP (100 nM) gradient was performed with dHL-60-EGFP-Myo1f cells or isolated human neutrophils. (A) Live cell imaging of the human neutrophil nucleus (labeled with Hoechst 5 µM, green) and the collagen meshwork (fire and red) using confocal reflection/fluorescence microscopy. Serial images of a movie of a representative neutrophil migrating through a meshwork of collagen fibers at indicated time points. Confocal reflection images demonstrate the architecture of the collagen gel as well as the neutrophil body (fire and red). Fluorescence images depict the shape of the nucleus during migration within a collagen gel (green). Merge (yellow) shows the reflection image (red) and the nucleus (green). Schematic outline of the shape of the cell (red) and the nucleus (green) while the neutrophil squeezes through a narrow pore. Arrows indicate the pore (white). Neutrophil localization is normalized to the position of the pore. Scale bar, 10 µm. The cell is representative for a total of 12 cells from 3 independent experiments. (B) Live cell imaging of a migrating dHL-60-EGFP-Myo1f cell using spinning-disk confocal microscopy. Pseudo-colored images demonstrate the morphology of the nucleus (gray) of dHL-60 EGFP-Myo1f cells, the subcellular localization of Actin (red) and Myo1f (green), as well as the colocalization of Actin and Myo1f (arrowheads). Arrows indicate constriction site. Scale bar, 10 µm. Color scales, heat map. Triangle indicates orientation of fMLP gradient. The cell is representative for a total of 9 cells from 3 independent experiments. (C) Schematic outline of the nuclear morphology (black), localization of Actin (red) and Myo1f (green), and colocalization of Actin and Myo1f (yellow) during 3D migration at indicated time points. The border of the cell body is depicted in gray.
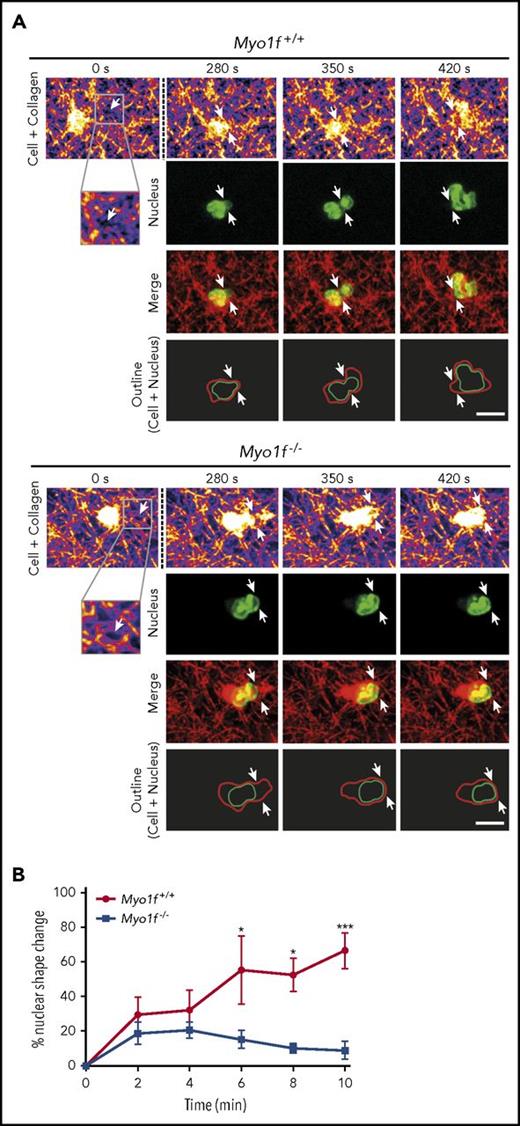
To study the functional impact of Myo1f during 3D migration in detail, we performed 3D migration toward a CXCL1 gradient using Hoechst-labeled murine Myo1f+/+ and Myo1f−/− neutrophils (Figure 6A-B). Real-time fluorescence microscopic images revealed that the nucleus of Myo1f+/+ neutrophils deformed during 3D migration as expected. Starting from a roundish, multilobular shaped nucleus (280 seconds), the nucleus elongated (350 seconds) to squeeze the cell body through the narrow pore (white arrows) and was refolded (420 seconds) into a spherical morphology (Figure 6A; supplemental Video 2). In sharp contrast, Myo1f-deficient neutrophils failed to deform their nuclei during 3D migration. The nucleus kept a rather spherical morphology without striking shape change during the entire recording time, suggesting that defective nucleus deformation was the reason for impaired 3D migration in the genetic absence of Myo1f. Quantitative analysis of nuclear morphology confirmed the shape change of nuclei of Myo1f+/+ neutrophils during 3D migration (Figure 6B). However, this change in nuclear morphology was almost absent in Myo1f−/− neutrophils, meaning that the shape change of the nucleus for successful 3D migration did not occur in the genetic absence of Myo1f. These results demonstrate that Myo1f-deficient neutrophils were almost unable to perform nucleus deformation resulting in compromised 3D migration through physical barriers in the meshwork of collagen fibers.
Myo1f−/−nuclei failed to deform during 3D migration.Myo1f+/+ and Myo1f−/− neutrophils migrating in a 1.5 mg/mL collagen gel toward a CXCL1 (100 ng/mL) gradient. (A) Live cell imaging of the neutrophil nucleus (labeled with Hoechst 5 µM, green) and the collagen meshwork (fire and red) using confocal reflection/fluorescence microscopy. Serial images of movies of representative neutrophils migrating through a meshwork of collagen fibers at indicated time points. Confocal reflection images demonstrate the architecture of the collagen gel as well as the neutrophil body (fire and red). Fluorescence images depict the shape of the nucleus during migration within a collagen gel (green). Merge (yellow) shows the reflection image (red) and the nucleus (green). Schematic outline of the shape of the cell (red) and the nucleus (green) while the neutrophils squeeze through narrow pores. Arrows indicate the pores (white). Neutrophil localization is normalized to the position of the pore. Scale bar, 10 µm. The cells are representative for a total of 9 cells from 3 independent experiments. (B) Percentage of nuclear shape change during neutrophil 3D migration at indicated time points based on individual nuclear shape of each neutrophil during the observation period of 30 minutes using spinning-disk confocal microscopy. Time point 0 minutes represents the most spherical nuclear morphology of each individual cell that was observed for 10 minutes thereafter. n = 3 independent experiments; mean ± SEM; *P < .05; ***P < .001 (2-way ANOVA, Sidak multiple comparison test).
Myo1f−/−nuclei failed to deform during 3D migration.Myo1f+/+ and Myo1f−/− neutrophils migrating in a 1.5 mg/mL collagen gel toward a CXCL1 (100 ng/mL) gradient. (A) Live cell imaging of the neutrophil nucleus (labeled with Hoechst 5 µM, green) and the collagen meshwork (fire and red) using confocal reflection/fluorescence microscopy. Serial images of movies of representative neutrophils migrating through a meshwork of collagen fibers at indicated time points. Confocal reflection images demonstrate the architecture of the collagen gel as well as the neutrophil body (fire and red). Fluorescence images depict the shape of the nucleus during migration within a collagen gel (green). Merge (yellow) shows the reflection image (red) and the nucleus (green). Schematic outline of the shape of the cell (red) and the nucleus (green) while the neutrophils squeeze through narrow pores. Arrows indicate the pores (white). Neutrophil localization is normalized to the position of the pore. Scale bar, 10 µm. The cells are representative for a total of 9 cells from 3 independent experiments. (B) Percentage of nuclear shape change during neutrophil 3D migration at indicated time points based on individual nuclear shape of each neutrophil during the observation period of 30 minutes using spinning-disk confocal microscopy. Time point 0 minutes represents the most spherical nuclear morphology of each individual cell that was observed for 10 minutes thereafter. n = 3 independent experiments; mean ± SEM; *P < .05; ***P < .001 (2-way ANOVA, Sidak multiple comparison test).
Discussion
In our study, we identified the unconventional class I Myo1f to be essential for the deformation of the nucleus during migration through restriction sites. Myo1f is therefore indispensable for neutrophil migration in confined 3D environments, that is, transmigration and migration in collagen networks. A previous study22 demonstrated that Myo1f−/− neutrophils exhibited an abnormally increased adhesion to the β2 integrin ligand rmICAM-1 and decreased 2D migration velocity under static conditions in vitro when cells were exposed to fibronectin and poly-L-lysine. Furthermore, it was observed that Myo1f−/− mice failed to control infection with Listeria monocytogenes and postulated that this defect was attributed to impaired mobility of neutrophils caused by increased adhesion. However, we found that mechanotaxis and chemotaxis in 2D environments were completely normal in the genetic absence of Myo1f indicating that the increased adhesion per se, which was also observed in the present study, did not simply hamper the mobility of neutrophils and cannot account for the accumulation defect in the tissue. To our knowledge nuclear deformability has no impact on adhesion under static conditions.
Interestingly, we detected increased intravascular neutrophil accumulation in the LPS-induced acute lung injury model in Myo1f−/− compared with Myo1f+/+ mice. The microvasculature of the lungs and the liver is characterized by low shear rates which may account for their unusual responses to inflammatory stimuli.36 Here, neutrophil recruitment does not uniformly follow the canonical cascade of adhesion and activation events known for other vascular beds.37,38 Thus, the local hemodynamic and microvascular features may contribute to the increased intravascular accumulation of Myo1f-deficient neutrophils in the lungs. In the present study, analysis of neutrophil adhesion in vitro under flow conditions (1 dyne/cm2) revealed normal adhesion of Myo1f−/− neutrophils compared with Myo1f+/+ neutrophils. Moreover, studying leukocyte rolling and adhesion by intravital microscopy in the cremaster muscle model using Myo1f+/+ and Myo1f−/− mice confirmed intact rolling and adhesion of Myo1f−/− neutrophils. Furthermore, spreading and polarization in 2D environments were unaffected in the presence of ICAM-1 or fibrinogen in Myo1f−/− neutrophils under flow conditions in vitro. This was in contrast to the above-mentioned study22 where neutrophil spreading was analyzed under static conditions and found to be increased in the absence of Myo1f, demonstrating the critical importance of the experimental conditions for the outcome of the experiments. Another study on Myo1f−/− macrophages revealed severely reduced spreading in the absence of Myo1f upon LPS stimulation.39 Thus, the biological function of Myo1f has evolved differently in diverse leukocyte subtypes. In summary, we conclude that Myo1f was dispensable for rolling, adhesion, polarization, spreading, and 2D migration under physiological flow conditions. Thus, the increased adhesion under static conditions did not represent the mechanism causing impaired neutrophil trafficking under physiological flow conditions. Here, analysis of different in vivo models of acute inflammation revealed a dramatic reduction in perivascular neutrophils in Myo1f−/− mice compared with Myo1f+/+ mice, which is in line with findings by Kim et al.22
To unravel the mechanism for the dramatic extravasation defect, we examined the role of Myo1f for neutrophil migration in confined 3D environments and identified Myo1f to be important for migration through restrictive pores. Previous studies showed that leukocyte transmigration and interstitial migration require extensive and rapid translocation and deformation of the nucleus to squeeze the cell through constrictions smaller than their diameter.14,16,40 The nucleus of human neutrophils is composed of 2 to 6 nuclear lobes with a diameter of 2 µm and a connection segment between 2 lobes with a size of ∼0.5 µm.41,42 In contrast to nuclei of human neutrophils, murine neutrophils tend to form ring-like nuclei with 1 to 2 lobes.43 Due to the low expression of LaminA/C in the nuclear envelope of both human and murine neutrophils, neutrophil nuclei are in general very flexible and elongate during transmigration.35,44 Here, we analyzed the morphology of the nucleus during migration in 3D environments and identified impaired nucleus deformation of Myo1f−/− neutrophils, compromising migration through narrow pores.
Interstitial migration not only depends on the overall cellular deformability but also on substrate porosity.34 In vivo interfibrillar spaces in tissues have been shown to range from 2 to 30 µm.45 Significant nuclear deformation has been shown to be required for neutrophil migration within tissues with pore cross sections of 2 to 20 µm2.34 Accordingly, we performed 3D migration experiments in gels with a collagen concentration of 3.0 mg/mL, yielding pore cross sections ranging between 2 and 3 µm2 and gels with a collagen concentration of 1.5 mg/mL resulting in pore cross sections of 10 to 12 µm2 as estimated in an earlier study.34 Thus, nuclear deformability and tissue porosity defined cell migration in the settings used. This was also true for transwell assays as a pore diameter of 3 µm (5 µm) corresponds to a cross-sectional area of 7.1 µm2 (19.6 µm2, A = π r2). In line with findings by Wolf et al,34 we demonstrated that neutrophil migration was reduced in high-density compared with low-density collagen gels. The genetic absence of Myo1f further compromised neutrophil migration through narrow pores with cross sections between 2 and 20 µm2 when compared with Myo1f+/+ neutrophils, suggesting an important role of Myo1f for nuclear deformation during migration in confined 3D environments.
The dynamic interactions between the cytoskeleton and the nucleus are an important prerequisite for cell locomotion and nucleus deformation during migration in 3D environments.12,15,16,46 The presented scheme in Figure 5C illustrates 3 different phases dHL-60 cells undergo during migration through constriction sites. In line with a previous study,35 the squeezing of the dHL-60 cell nucleus was initiated by forming a nuclear lobe followed by the elongation of the nucleus to squeeze the cell body through the narrow pore. Similar phases were observed in primary human neutrophils. Moreover, we identified Myo1f to be enriched at the back and the front of the elongated nucleus during the initiation and deformation phases and speculated that Myo1f was involved in pushing and/or pulling the nucleus through the constriction sites. Moreover, we observed a colocalization of Myo1f and Actin at the rear and the front of the migrating cell as well as at the constriction sites, suggesting that Myo1f was required for driving the nuclear deformation during migration through narrow pores.
In addition to the high malleability of the neutrophil nucleus, the nucleus-cytoskeleton connection is essential for 3D migration by transmitting force from the cytoskeleton to the inside of the nucleus.47 The linkers of nucleocytoskeleton to cytoskeleton (LINC) complexes are proteins spanning the nuclear membrane and regulating actin bundles attached to the nucleus during 3D migration in mouse embryonic fibroblasts.48-51 However, previous studies using primary human neutrophils and dHL-60 cells showed that the lobulated neutrophil nucleus is characterized by low expression of LINC complex proteins providing a flexible nuclear envelope structure resulting in a malleable cell.52-54 Thus, it seems reasonable to speculate that Myo1f may link the cytoskeleton to the nuclear envelope via its TH1 domain to provide high malleability of the neutrophil nucleus. However, the exact mechanism by which Myo1f causes nuclear deformation during 3D migration of neutrophils remains elusive and requires further analysis. In summary, Myo1f represents a novel and important molecular player specifically required for neutrophil migration in 3D environments by regulating the dynamic deformation of the nucleus during migration through physical barriers. By coordinating this complex process, Myo1f is indispensable for neutrophil trafficking in innate immunity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jennifer Truong and Yvonne Jansen for excellent technical assistance as well as Andreas Thomae and Steffen Dietzel (Core Facility Bioimaging, BioMedical Center, Ludwig-Maximilians-Universität München) for their help with confocal reflection fluorescence microscopy.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 914, project A02 [B.W.], B01 [M. Sperandio], B08 [O.S.], Z03 [B.W. and M. Sperandio]), the Else Kröner-Fresenius-Stiftung (grant 2014_A217) (B.W.), and the FöFoLe program of the Ludwig-Maximilians-Universität München (B.W.).
Authorship
Contribution: M. Salvermoser designed and carried out research, analyzed data, and wrote the manuscript; R.P. performed spinning-disk confocal microscopy experiments, analyzed data, and wrote the manuscript; intravital microscopy was performed by L.T.W. together with M. Sperandio; P.L. and A.Z. generated and characterized the HL-60 EGFP-Myo1f cell line; M.D. and O.S. carried out the acute lung injury experiments and analyzed the data; and B.W. designed the overall study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara Walzog, Walter Brendel Centre of Experimental Medicine, University Hospital, Institute of Cardiovascular Physiology and Pathophysiology, Biomedical Center, LMU Munich, Planegg-Martinsried, Germany; e-mail: walzog@lrz.uni-muenchen.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal