Abstract
Background: Outcomes for patients with diffuse large B-cell lymphoma (DLBCL) remain heterogeneous. Several clinical and molecular risk factors have been described, both at time of diagnosis (i.e., International Prognostic Index and cell of origin) and during therapy (i.e., interim PET scans). However, these risk factors fail to consistently identify individuals destined for treatment failure. We recently described circulating tumor DNA (ctDNA) as a novel biomarker in DLBCL, demonstrating pretreatment ctDNA levels are prognostic of outcomes (Scherer F, Sci Transl Med 2016). A major advantage of ctDNA assessment is the ability to collect serial assessments over time. However, the best approach for integrating multiple ctDNA measurements with established prognostic factors remains unknown. Using the dynamics of ctDNA, we developed a method to quantify personalized disease risk as it changes during therapy.
Methods: We profiled 468 samples from 125 subjects collected during their first three cycles of immunochemotherapy using cancer personalized profiling by deep sequencing (CAPP-Seq). Early ctDNA dynamics were correlated with outcomes in a training cohort to define a response threshold that was tested in a validation cohort. Finally, we integrated ctDNA and clinical factors to dynamically assess disease risk over time in a continuous model.
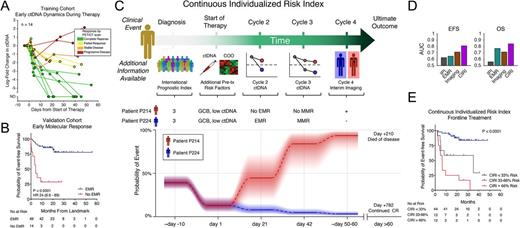
Results: Prior to therapy, ctDNA was detectable in 98% of subjects. Pretreatment levels of ctDNA were prognostic of event-free survival in patients receiving either frontline anthracycline-based (n=92) or salvage regimens (n=33) (EFS: HR 2.7, 95% CI 1.5-4.7, P=0.0005), confirming prior results. In the training cohort, ctDNA levels changed rapidly, with a 2-log or 100-fold decrease after one cycle (early molecular response, EMR) stratifying outcomes (Fig 1A). In the validation cohort, patients achieving EMR had superior outcomes at 24-months to those who did not (Fig 1B) (EFS: HR 24, 95% CI 6.6-89, P<0.0001). These results remained significant in subgroups of patients receiving either frontline or salvage therapy. Circulating tumor DNA levels continued to decline during cycle 2, such that a distinct threshold after two cycles could also be trained. After two cycles, a 2.5-log decrease (major molecular response, MMR) also stratified patients for EFS (HR: 8.6, 95% CI 2.2-33, P=0.002).
Next, we integrated serial ctDNA measurements with established risk-factors to develop a model to predict an individual's disease risk. This model - the Continuous Individualized Risk Index (CIRI) - provides a personalized estimate of disease risk over time. As more information becomes available during a patient's course of disease, CIRI updates the disease risk, integrating the new information (Fig 1C). In patients receiving frontline therapy, CIRI outperformed the IPI for identification of 24-month EFS and OS (Fig 1D-E) (EFS24: AUC 0.64 vs 0.79; net reclassification improvement 0.47, P=0.02; OS: AUC 0.56 vs 0.84; net reclassification improvement 0.74, P=0.004).
Conclusions: Baseline and interim ctDNA measurements have prognostic significance in aggressive lymphomas. Integration of serial ctDNA measurements through a continuous, dynamic risk model can identify personalized outcome probabilities, yielding superior risk estimates. Dynamic risk assessment is potentially widely applicable and could guide future personalized therapeutic approaches.
Figure 1: A) A spider-plot depicts the dynamics of ctDNA during the first two cycles of therapy in 14 subjects. B) A Kaplan-Meier estimate depicts EFS for patients achieving or not achieving EMR. C) A schema for CIRI is shown. When a patient is diagnosed with DLBCL, the IPI is calculated, giving an initial estimate of risk. Additional pretreatment risk factors (i.e., cell of origin and pretreatment ctDNA) can be added, thereby updating the estimate of risk. As a patient undergoes therapy, further predictors of risk are obtained, such as ctDNA at cycle 2 and cycle 3, and interim imaging. These predictors can be used to update the patient's estimated risk. Below is a CIRI risk model for two exemplar patients with the same initial pretreatment risk factors. Patient P214 (red) died of disease at day 210, while patient P224 (blue) is in a continued complete response. D) AUC for prediction of EFS and OS by IPI, EMR, interim PET scans, and CIRI. E) A Kaplan-Meier estimate demonstrates risk stratification of EFS by CIRI.
Advani: Sutro: Consultancy; Cell Medica: Research Funding; Janssen: Research Funding; Bristol-Myers Squibb: Consultancy, Research Funding; Celgene: Research Funding; Nanostring: Consultancy; Bayer Healthcare Pharmaceuticals: Research Funding; Juno Therapeutics: Consultancy; FortySeven: Research Funding; Gilead: Consultancy; Spectrum: Consultancy; Pharmacyclics: Research Funding; Agensys: Research Funding; Regeneron: Research Funding; Millennium: Research Funding; Infinity: Research Funding; Merck: Research Funding; Kura: Research Funding; Pharmacyclics: Consultancy; Seattle Genetics: Research Funding; Genentech: Research Funding. Newman: Roche: Consultancy. Westin: Celgene: Membership on an entity's Board of Directors or advisory committees; Apotex: Membership on an entity's Board of Directors or advisory committees; Kite Pharma: Membership on an entity's Board of Directors or advisory committees; Novartis Pharmaceuticals Corporation: Membership on an entity's Board of Directors or advisory committees. Gaidano: Janssen: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Gilead: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria. Diehn: Varian Medical Systems: Research Funding; Roche: Consultancy; Novartis: Consultancy; Quanticel Pharmaceuticals: Consultancy. Alizadeh: Genentech: Consultancy; Celgene: Consultancy; CiberMed: Consultancy; Roche: Consultancy; Gilead: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal