Abstract
INTRODUCTION
Next-generation sequencing of diffuse large B cell lymphoma (DLBCL) has revealed a vast landscape of genetic alterations. Unfortunately, very few of these carry a significant prognostic power or confer a different response to therapy. Unique, in this respect, are mutations in TP53, which have been implicated in over a third of the refractory cases to frontline regimes. This study aimed to evaluate the association between clinical phenotype, disease course and specific genomic alterations of TP53mut DLBCL, comparing de novo to relapsed or refractory disease (R/R), and chemo-sensitive to chemo-resistant cases.
METHODS
We performed a retrospective analysis of patients with DLBCL, treated at Memorial Sloan Kettering Cancer Center, harboring a TP53mut. Formalin fixed, paraffin embedded samples were sequenced using a custom DNA capture panel. Germline variants were filtered against the 1000 Genomes Project. Significant non-synonymous variants were identified by mapping to the COSMIC database, as well as clear inactivating mutations in established tumor suppressor genes. Analysis for TP53 mutations was performed per their affected p53 protein domains, including DNA binding motifs (Loop-L2, Loop-L3, Loop-Sheet-Helix), evolutionary conserved regions, non-conserved and truncating non-missense mutations.1Sequencing was performed at various time-point in the course of the disease. We evaluated TP53mut allele frequency (AF), overall number of TP53mut /patient, and concurrent alterations in other genes. Disease characteristics and clinical course were extracted for first line treatment of DLBCL, as well as from R/R cases. Cell of origin (COO) determination and MYC, BCL2 and BCL6 rearrangements at the time of sequencing were recorded. We evaluated response to treatment and survival in the subset of patients sequenced at diagnosis. Transformed indolent lymphoma (t-DLBCL) cases were evaluated in respects to the time of transformation, but considered R/R if previously treated with chemotherapy.
RESULTS
Of 360 sequenced DLBCL cases, 84 were found to carry a TP53 mutation (35 de novo ; 49 R/R). Of these, 61% had a stage IV disease at presentation, involving extra-nodal sites (mostly musculoskeletal, lung, gastrointestinal and hepatic). Central nervous system involvement was rare throughout the course of the disease (2%). t-DLBCL represented a considerable part of the TP53mut population, more so in R/R disease (59% vs. 29%, p=0.01), where it originated more commonly from follicular lymphoma (76% vs. 50%, p=0.2). R/R was also enriched for Double-Hit disease (41% vs. 20%, p=0.06) and for MYC copy number alterations (59% vs. 20%, p=0.02), though the number of evaluable cases was small.
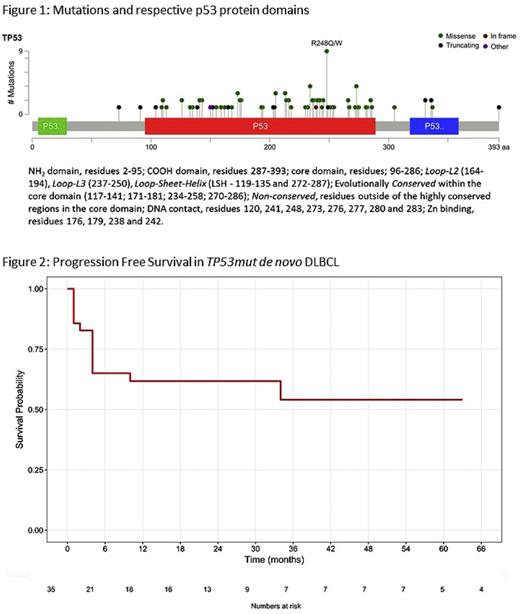
We identified 105 TP53 mutations, with most patients (86%) harboring only one. Overall, 49% of patients had a mutation involving a DNA binding motif, 26% a non-missense truncating mutations, and 35% a mutation affecting non- DNA binding motif regions (figure 1). R/R disease was associated with a higher AF (0.52 vs. 0.35, p=0.01), but not with a higher overall number of TP53mut or the affected p53 domain. Patients had a median of 4 concurrent mutations in other genes without a clear association with R/R .
Among the R/R patients, 65% were refractory to their first-line chemotherapy, with a median of 4 therapy lines overall. Further, of the 23 patients receiving autologous stem cell transplantation, 52% were refractory. Taken together, these observations suggest that TP53mut may be an early event conferring a high rate of multi-drug resistance. Refractoriness to frontline therapy was also observed in 49% of the de novo case. However, with a median follow-up of 30 months, 2y PFS was nearly 50%, demonstrating a subset of TP53mut patients who fare well (figure 2). There was no association between PFS and the site of TP53mut, total number of TP53 mutations or number and type of concurrent mutations.
CONCLUSIONS
TP53mut is associated with multi-drug resistance and a worse prognosis. However, nearly 50% of newly diagnosed patients can be expected to respond to frontline regimens, many of whom achieving long term remissions. We could not find a clear genomic feature to differentiate Responsive from Refractory patients. Future research should focus on prospectively examining the role of TP53mut in predicting treatment outcomes.
Zelenetz: Amgen: Consultancy; Celgene: Consultancy. Moskowitz: Celgene: Consultancy; Pharmacyclics: Research Funding; Merck: Consultancy, Research Funding; Seattle Genetics: Consultancy, Other: Ad Board, Research Funding; Genentech BioOncology: Consultancy. Palomba: Merck: Consultancy. Noy: Pharmacyclics LLC, an AbbVie Company: Honoraria, Other: Travel, Accommodation, Expenses, Research Funding, Speakers Bureau. Straus: Received consulting fee from Seattle Genetics for involvement in the research: Consultancy. Gerecitano: Bayer: Consultancy, Membership on an entity's Board of Directors or advisory committees; Royal Bank of Canada: Honoraria, Membership on an entity's Board of Directors or advisory committees; Abbvie: Honoraria, Membership on an entity's Board of Directors or advisory committees; Incyte: Consultancy, Membership on an entity's Board of Directors or advisory committees; Aratana: Consultancy, Membership on an entity's Board of Directors or advisory committees; Arcus Medica: Consultancy, Membership on an entity's Board of Directors or advisory committees; Gilead: Honoraria, Membership on an entity's Board of Directors or advisory committees; Orexo: Honoraria, Membership on an entity's Board of Directors or advisory committees; Samus Therapeutics: Honoraria, Membership on an entity's Board of Directors or advisory committees; Genentech: Honoraria, Membership on an entity's Board of Directors or advisory committees; Merck: Honoraria, Membership on an entity's Board of Directors or advisory committees; Mass Medical International: Honoraria, Membership on an entity's Board of Directors or advisory committees. Horwitz: Kyowa Hakko Kirin: Consultancy, Research Funding; Infinity/Verastem: Consultancy, Research Funding; Forty-Seven: Consultancy, Research Funding; Mundipharma: Consultancy; HUYA: Consultancy; BMS: Consultancy; Seattle Genetics: Consultancy, Research Funding; Spectrum Pharmaceuticals: Research Funding; Aileron Therapeutics: Research Funding; Millennium Pharmaceuticals, Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited: Research Funding; Millennium Pharmaceuticals, Inc.: Consultancy; Celgene: Consultancy, Research Funding; ADCT Therapeutics: Research Funding. Moskowitz: ADC Therapeutics: Research Funding; Seattle Genetics: Honoraria, Research Funding; Incyte: Research Funding; Bristol Myers-Squibb: Consultancy, Research Funding; Takeda: Honoraria. Hamlin: Seattle Geneitcs: Other: research support; Celgene: Consultancy, Honoraria; Novartis: Other: research support; Portola: Consultancy, Honoraria, Other: research support; Gilead: Consultancy, Honoraria; Incyte: Other: research support. Kumar: Seattle Genetics: Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees. He: Foundation Medicine, Inc: Employment, Other: Stock. Miller: Foundation Medicine: Employment, Other: Stock. Dogan: Peer Review Institute: Consultancy; Celgene: Consultancy; Seattle Genetics: Consultancy, Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees; Roche Pharmaceuticals: Consultancy. Younes: Bayer: Honoraria; Merck: Honoraria; Janssen: Honoraria; Sanofi: Honoraria; Incyte: Honoraria; Celgene: Honoraria; Bristol-Myers Squibb: Honoraria; Takeda Millenium: Honoraria; Johnson & Johnson: Research Funding; Seattle Genetics: Honoraria; Novartis: Research Funding; Curis: Research Funding; Roche: Consultancy, Honoraria, Other: Third-party medical writing assistance, under the direction of Anas Younes, was provided by Scott Malkin of Gardiner-Caldwell Communications, and was funded by F. Hoffmann-La Roche Ltd.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal