Abstract
When considering HLA-matched hematopoietic cell transplantation (HCT), sibling and unrelated donors (UDs) are biologically different because UD-HCT is typically performed across HLA-DP disparities absent in sibling HCT. Mismatched HLA-DP is targeted by direct alloreactive T cell responses with important implications for graft-versus-host disease and graft-versus-leukemia. This concise review details special features of HLA-DP as model antigens for clinically permissive mismatches mediating limited T-cell alloreactivity with minimal toxicity, and describes future avenues for their exploitation in cellular immunotherapy of malignant blood disorders.
Introduction
The importance of allele-level HLA-A, -B, -C, -DRB1 donor-recipient matching for the clinical success of allogeneic hematopoietic cell transplantation (allo-HCT) is well established.1-4 Biologically, alloreactive donor T-cell responses to mismatched HLA antigens of the host lead to deleterious graft-versus-host disease (GVHD) on the one hand and favorable graft-versus-leukemia (GVL) on the other.5-8 Dissecting GVHD from GVL is the holy grail of successful allo-HCT for the treatment of malignant blood disorders. To achieve this ambitious goal, it is crucial to understand the molecular target structures recognized by alloreactive donor T cells mediating these effects.9,10 These target structures are fundamentally different in allo-HCT from HLA-matched sibling and unrelated donors (UDs), which are often erroneously considered as a single entity (“HLA-matched” HCT). However, in sibling HCT the targets of T-cell alloreactivity are almost exclusively minor histocompatibility antigens (mHAgs),11-13 whereas most HLA-matched UDs additionally present mismatches for the HLA-DP antigens encoded in the major histocompatibility complex (MHC).14,15 The latter can elicit direct alloreactive T-cell responses with important implications for both GVHD and GVL.16 Reflecting distinct features of its polymorphism, HLA-DPB1 has been the first locus to be explored as a model for clinically permissive, that is, well tolerated, donor-recipient HLA mismatches. More recently, new strategies for exploiting mismatched HLA-DP in adoptive cellular immunotherapy of malignant blood disorders are emerging. These developments have prompted increasing interest in HLA-DP as a challenge and opportunity in HLA-matched UD-HCT.
Molecular targets of GVHD and GVL after HLA-matched HCT
mHAgs in HLA-matched sibling HCT
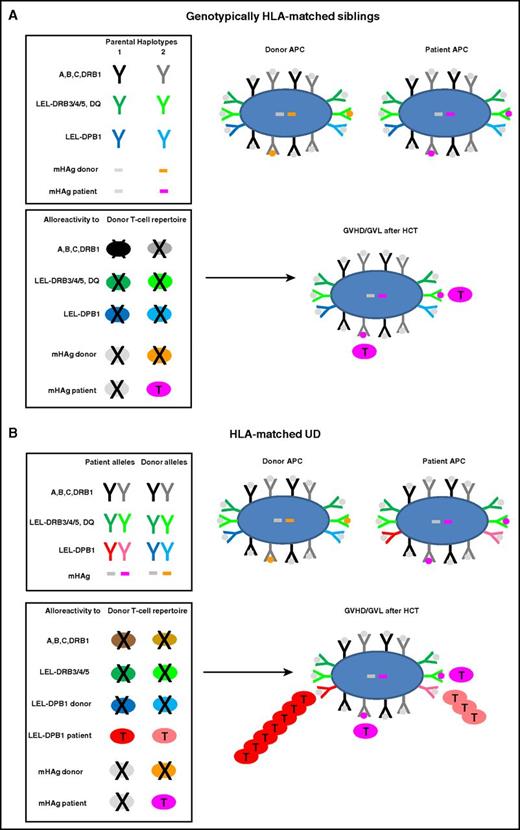
Alloreactive donor T cells play an important role in post-HCT GVHD and GVL, a complex process reviewed in detail elsewhere.8,17-20 In HLA-matched sibling HCT, T-cell alloreactivity is directed against mHAgs, that is, polymorphic non-self-peptides encoded anywhere in the human genome.9,11-13 Allorecognition of mHAgs is mediated by conventional self-HLA–restricted, foreign peptide-specific T cells whose repertoire is shaped by positive and negative selection in the thymus.21,22 Because allogeneic mHAgs are not expressed by thymic antigen-presenting cells (APCs) of self-origin, alloreactive T cells specific for mHAgs occur almost exclusively in the naive T-cell repertoire.16 Recent clinical protocols of selective depletion of naive T cells from the graft23,24 therefore have the potential to profoundly ablate T-cell alloreactivity after HLA-matched sibling HCT. This approach is promising for nonmalignant disorders where GVL is not an issue, but must be carefully weighed against the disease risk for individual patients transplanted for malignant disease. The exclusive peptide-specificity of mHAg-specific alloreactive T cells accounts for their low precursor frequency of <0.1%, explaining why not only GVHD but also GVL is limited after HLA-matched sibling HCT.24-26 Genotypically HLA-matched sibling donors generally share both alleles of HLA-A, -B, -C, -DR, -DQ, and -DP (12 of 12) because they have inherited the same parental copies of chromosome 6 encompassing the MHC (Figure 1A). Exceptions do occur in 1% to 5% of cases, accounted for by genomic recombination, with the highest frequency reported for HLA-DP due to at least 1 recombination hotspot between DP and DQ.27-29
Molecular targets of GVHD and GVL after HCT from HLA-matched siblings or UD. HLA molecules and mHAg peptides are depicted as Y or bars, and APCs and T cells as large or small oval shapes, respectively. T cells that undergo positive or negative selection in the thymus are labeled with a T or an X, respectively. Self-peptides presented by HLA molecules on the surface of an APC are indicated with gray dots; mHAg peptides are indicated with orange or pink dots. (A) Genotypically HLA-matched siblings. In the left top box, the 2 parental HLA haplotypes are schematically represented, along with 2 examples of mHAgs of which 1 is shared (gray bars) and the other is not shared (orange and pink bars) by the 2 siblings. In the bottom box, the T-cell repertoire of the sibling donor after thymic education is shown. Self-reactive T cells against HLA molecules have been clonally deleted by negative selection, whereas T cells alloreactive to the unshared mHAg (pink T cells) have undergone positive selection and mediate GVHD and/or GVL after HLA-matched sibling HCT. (B) HLA-matched UD. The top box shows the HLA component of a typical 10 of 10 UD-recipient pair matched for both HLA-A, -B, -C-DRB1 alleles (in black and gray) as well as for the LEL-DRB3/4/5 and -DQ (in dark and light green), but mismatched for both LEL-DPB1 alleles (in dark and light blue and red, respectively). Moreover, in this example, patient and donor are mismatched for the same mHAg as in panel A. The bottom box shows the donor T-cell repertoire after positive and negative selection, which contains T cells alloreactive not only to mismatched mHAgs but also to both mismatched LEL-DPB1 alleles. In this example, 1 of the 2 mismatched HLA-DPB1 alleles is nonpermissive (dark red) whereas the other one is permissive (light red), giving rise to direct T-cell responses of different magnitude but generally stronger than the response to mHAg mismatches. Together, these alloreactive T cells mediate GVHD and/or GVL after HLA-matched UD-HCT.
Molecular targets of GVHD and GVL after HCT from HLA-matched siblings or UD. HLA molecules and mHAg peptides are depicted as Y or bars, and APCs and T cells as large or small oval shapes, respectively. T cells that undergo positive or negative selection in the thymus are labeled with a T or an X, respectively. Self-peptides presented by HLA molecules on the surface of an APC are indicated with gray dots; mHAg peptides are indicated with orange or pink dots. (A) Genotypically HLA-matched siblings. In the left top box, the 2 parental HLA haplotypes are schematically represented, along with 2 examples of mHAgs of which 1 is shared (gray bars) and the other is not shared (orange and pink bars) by the 2 siblings. In the bottom box, the T-cell repertoire of the sibling donor after thymic education is shown. Self-reactive T cells against HLA molecules have been clonally deleted by negative selection, whereas T cells alloreactive to the unshared mHAg (pink T cells) have undergone positive selection and mediate GVHD and/or GVL after HLA-matched sibling HCT. (B) HLA-matched UD. The top box shows the HLA component of a typical 10 of 10 UD-recipient pair matched for both HLA-A, -B, -C-DRB1 alleles (in black and gray) as well as for the LEL-DRB3/4/5 and -DQ (in dark and light green), but mismatched for both LEL-DPB1 alleles (in dark and light blue and red, respectively). Moreover, in this example, patient and donor are mismatched for the same mHAg as in panel A. The bottom box shows the donor T-cell repertoire after positive and negative selection, which contains T cells alloreactive not only to mismatched mHAgs but also to both mismatched LEL-DPB1 alleles. In this example, 1 of the 2 mismatched HLA-DPB1 alleles is nonpermissive (dark red) whereas the other one is permissive (light red), giving rise to direct T-cell responses of different magnitude but generally stronger than the response to mHAg mismatches. Together, these alloreactive T cells mediate GVHD and/or GVL after HLA-matched UD-HCT.
HLA-DP in HLA-matched UD-HCT
In UD-HCT, patients and their HLA-matched donors carry the same 8 of 8 alleles at HLA-A, -B, -C, and -DRB1, without sharing their respective genetic backgrounds (Figure 1B). It has been consistently shown that single-nucleotide-polymorphism (SNP) variability within the MHC is higher in unrelated compared with sibling donors.30 Of note, genetic disparity between unrelated individuals frequently includes the HLA-DRB3/4/5, -DQ, and -DP loci which are targets of direct T-cell alloreactivity, that is, cross-recognition by T cells specific for different self-HLA–restricted peptides regardless of their derivation from novel or recall antigens.9,10,31 This phenomenon explains why T cells mediating direct alloreactivity are found both in the naive and memory repertoire and have a surprisingly high precursor frequency of at least 1%.10,24,26,32 HLA-DRB3/4/5, -DQ, and -DP have relatively low expression levels and are therefore referred to as low-expression loci (LEL). Although recipient-donor disparity for individual LEL has a limited impact on the clinical outcome of HCT, combined LEL mismatches were shown to be negatively associated with survival after 7 of 8–matched UD-HCT.33 Moreover, alloreactive T cells targeted to LEL have been recovered from patients with GVHD and GVL.34-37 Due to strong linkage disequilibrium (LD) between HLA-DRB1 and -DRB3/4/5 and -DQ, mismatches for these LEL are relatively rare in UD-HCT. In contrast, HLA-DP mismatches are present in over 80% of HLA-matched UD-HCT, reflecting their low LD with the other class II loci.38,39 T-cell alloreactivity targeted to mismatched HLA-DP is therefore a hallmark of HLA-matched UD-HCT, in contrast to HLA-matched sibling HCT, with important implications for GVHD and GVL as discussed in “Clinical relevance of HLA-DPB1 disparity in unrelated HCT.”
Structure and polymorphism of HLA-DP
HLA-DP class II αβ-chain heterodimers are constitutively expressed on different immune cells including professional APCs, and can be induced on nonhematopoietic tissues under inflammatory conditions.40 Importantly, HLA-DR+ leukemias and lymphomas generally coexpress HLA-DP, making it an attractive GVL target after allo-HCT.41 The HLA-DP-αβ chains are encoded by the DPA1 and DPB1 loci at the centromeric end of the MHC, at a distance of about 400 Mb from DQB1 from which they are separated by at least 1 recombination hotspot.27,42 In contrast to the weak LD between DP and the other HLA class II loci, LD for SNPs within DPA1 and DPB1 and between the 2 loci is strong, and shuffling of 6 hypervariable SNP regions in DPB1 accounts for most of its genomic variability. The reported polymorphism is significantly higher for DPB1 than for DPA1, with 894 and 53 alleles, respectively.43 Of note, only 12 DPB1 and 2 DPA1 alleles account for over 90% of the variants observed in worldwide populations.44 Therefore, homozygosity (ie, the same DP allele present on both chromosomes of an individual) is frequent, often resulting in unidirectional (ie, only in graft-versus-host [GVH] or host-versus-graft [HVG] direction) HLA-DP mismatches in UD-recipient pairs.
Clinical relevance of HLA-DPB1 disparity in unrelated HCT
HLA-DPB1 allele mismatches
HLA-DP was first discovered as a target of alloreactivity in primed lymphocyte testing.45 Initial evidence for a role of HLA-DPB1 in UD-HCT has been reported since the mid-1990s,34,35,46-48 and a large body of literature has accumulated to date (Table 1). Experimentally, the ability of a wide range of HLA-DPB1 allele mismatches to elicit polyclonal alloreactive T-cell responses has been confirmed.49 In line with these observations, HLA-DPB1 mismatches have been shown to be associated with increased risks of acute GVHD after 8 of 8 HLA-matched unrelated HCT.34,35,48 Importantly, however, they also mediate a significantly reduced risk of leukemia relapse, resulting in no net impact on overall survival.38,47,50-52 The protective effect of HLA-DPB1 allele mismatches against relapse is a clinical confirmation of their above-mentioned attractiveness as GVL targets. This notion is further substantiated by elegant experimental work showing that T cells alloreactive to mismatched recipient HLA-DP can be recovered from patients who clear residual malignant disease in response to CD4+ donor lymphocyte infusions after UD-HCT.53,54 Finally, HLA-DP mismatches also have a well-recognized role in both cellular and humoral stem cell allograft rejection.46,55-58
Major milestone publications impacting practice in the clinical consideration of HLA-DPB1
| Reference . | No. . | Patient and HCT characteristics . | Era . | HLA matching* and clinical outcomes . | Major findings/impact . |
|---|---|---|---|---|---|
| 46 | 1 | CML | 2001 | 11/12 in GVH direction; case report, acute rejection | First evidence that mismatched HLA-DPB1 can be the target of CD4+ T-cell–mediated allograft rejection |
| Case report | |||||
| 47 | 143 | Various diseases | 1996-2001 | 10/10; HLA-DPB1 allele mismatches; increased aGVHD, decreased relapse | First evidence of a GVL effect targeted to HLA-DPB1 allele mismatches after UD-HCT |
| Myeloablative conditioning | |||||
| T-cell depletion | |||||
| 62 | 118 | Various diseases | 1995-2002 | 10/10; HLA-DPB1 nonpermissive TCE mismatches; increased aGVHD and NRM | First description of a T-cell epitope matching score for HLA-DPB1 |
| Myeloablative conditioning | |||||
| T-cell depletion | |||||
| 50 | 423 | Various diseases | 1996-2003 | 10/10; HLA-DPB1 allele mismatches; increased aGVHD, decreased relapse, better OS in ALL | Confirmation of Shaw et al47 and possibly improved survival of ALL patients after HLA-DPB1 mismatched HCT |
| Myeloablative conditioning | |||||
| T-cell depletion | |||||
| 55 | 72 | Thalassemia | 1992-2004 | 10/10; HLA-DPB1 nonpermissive HVG mismatches; increased graft rejection | First association of nonpermissive TCE mismatches with outcome of HCT for nonmalignant disease |
| 79 | N/A | N/A | N/A | N/A | First protocol of targeted epitope-specific HLA-DPB1 typing for UD-HCT |
| 38 | 5929 | Various diseases | 1984-2005 | 5-8/8; HLA-DPB1 allele mismatches; increased aGVHD, decreased relapse | First definitive confirmation of GVL targeted to HLA-DPB1 allele mismatches after UD-HCT in a T-cell–replete cohort |
| Registry study (14th IHIW) | |||||
| 53 | 1 | Lymphoblastic lymphoma | 2006 | 10/10; 1 permissive and 1 nonpermissive HlA-DPB1 TCE mismatch | First isolation of CD4+ T cells alloreactive to mismatched HLA-DPB1 from a patient clearing residual malignant disease after donor lymphocyte infusion from an UD |
| Case report | |||||
| 63 | 621 | Various diseases | 1999-2006 | 10/10; HLA-DPB1 nonpermissive TCE mismatches; increased aGVHD and NRM, worse OS | First demonstration of a negative impact of nonpermissive HLA-DPB1 mismatches in a large multicenter registry cohort |
| Myeloablative and reduced-intensity conditioning | |||||
| Registry study (IBMDR) | |||||
| 51 | 488 | AML, ALL, CML | 1996-2006 | 10/10 and <10/10; HLA-DPB1 allele mismatches; decreased relapse, increased NRM, worse OS in early disease in 10/10; improved OS in late disease in <10/10 | Differential impact of HLA-DPB1 allele mismatches in 10/10 or <10/10 according to disease status |
| Myeloablative and reduced-intensity conditioning | |||||
| T-cell depletion | |||||
| 49 | 4 | Healthy donors | N/A | In vitro generation of 16 T-cell clones alloreactive to a variety of HLA-DP specificities by stimulation with transfected HELA cells | Confirmation that CD4+ T-cell alloreactivity to HLA-DPB1 can be elicited by both permissive and nonpermissive mismatch combinations |
| 65 | 24 | Healthy donors | N/A | In vitro mixed lymphocyte reactions between 10/10 matched, HLA-DPB1 permissive or nonpermissive UD | First experimental evidence for more potent T-cell alloreactivity to nonpermissive than to permissive HLA-DPB1 TCE mismatches |
| 56 | 115 | AML, ALL, CML, MDS, UD-HCT | 1990-2002 | 6/12 to 12/12; retrospective matched control study graft failure vs engraftment | Graft failure is associated with donor-specific HLA antibodies, over half of them against HLA-DP |
| 57 | 592 | AML, ALL, MDS and others, UD-HCT | 2005-2009 | 9/12 to 12/12; prospective HLA antibody testing | Donor-specific antibodies in 1.4% of patients, all against mismatched HLA-DP; significantly associated with graft failure |
| 39 | 8539 | AML, ALL, CML, MDS | 1993-2007 | 10/10 and 9/10; HLA-DPB1 nonpermissive TCE mismatches; increased aGVHD and NRM, worse OS | First definitive demonstration of a negative impact of nonpermissive HLA-DPB1 mismatches on OS in 10/10 and 9/10 UD-HCT |
| Registry study (15th IHIW) | |||||
| 54 | 24 | Various diseases | 2000-2008 | 10/10; CD4+ DLI for mixed chimerism or relapse; ex vivo analysis of T cells alloreactive to HLA-DPB1 in responding or nonresponding patients | First demonstration of reliable emergence of CD4+ T cells alloreactive to HLA-DP in patients responding to DLI after UD-HCT |
| T-cell–depleted UD-HCT | |||||
| 68 | N/A | N/A | N/A | N/A | Launch of the first free online tool for HLA-DPB1 nonpermissive/permissive TCE match assignment |
| 36 | 2 | AML | N/A | 10/10; HLA-DPB1 nonpermissive GVH mismatch in both patients; severe aGVHD of the gut after CMV reactivation | Host-derived CMV-specific T cells can trigger CD4+ T-cell alloreactivity against mismatched HLA-DPB1 after UD-HCT, leading to severe GVHD |
| 2 case reports | |||||
| 66 | 1281 | AML, ALL, CML, MDS | 1988-2003 | 10/10; HLA-DPB1 nonpermissive GVH mismatches significantly lower relapse than permissive mismatches; no impact of concomitant mismatching for DPA1 | GVL effect by nonpermissive GVH mismatches; no impact of DPA1 on nonpermissive TCE mismatches |
| Registry study (CIBMTR) | |||||
| 4 | 8003 | AML, ALL, CML, MDS | 1999-2011 | 10/10 and 9/10; HLA-DPB1 nonpermissive TCE mismatches; increased NRM, worse OS in 10/10 but not in 9/10 | First confirmation of a negative impact of nonpermissive HLA-DPB1 mismatches in 10/10 on OS in an independent validation cohort from a registry |
| Registry study (CIBMTR) | |||||
| 71 | 4 | Healthy donors | N/A | Generation of 12 site-directed mutants of HLA-DPB1*09:01 at 10 key amino acid positions; investigation of their allorecognition by 17 T-cell effectors alloreactive to wild-type HLA-DPB1*09:01; determination of the median impact of each substitution on T-cell alloreactivity (FD) | Development of a FD score system for individual amino acid polymorphism in HLA-DPB1, and for HLA-DPB1 alleles. The FD score of any current or future HLA-DPB1 allele can be used to predict its assignment to TCE groups |
| 52 | 7898 | Various diseases | 1993-2010 | 10/10; role of HLA-locus mismatches including HLA-DPB1 allelic disparity; increased aGVHD, decreased relapse | Confirmation of lower relapse risk associated with HLA-DPB1 allele mismatches; no such association was observed with HLA-DRB1 allele mismatches |
| Registry study (JMDP) | |||||
| 64 | 2029 | AML, ALL, CML, MDS | 1988-2008 | 10/10; truly unidirectional GVH HLA-DPB1 expression mismatches; increased aGVHD when patient carries high-expression mismatch | First demonstration that expression levels determine nonpermissive GVH HLA-DPB1 mismatches; conclusive association between 3′ UTR expression SNPs and exon HLA-DPB1 types |
| Registry Study (CIBMTR) | |||||
| 80 | 1342 | Various diseases | 2000-2008 | 10/10; increased grade III-IV aGVHD with 2 allelic HLA-DPB1 mismatches, increased relapse with TCE nonpermissive HVG disparities | HLA-DPB1 impacts aGVHD and relapse by number of allelic mismatches and TCE HVG disparities, respectively |
| Registry study (RFGM) | |||||
| 69 | N/A | N/A | 2013 | N/A | Description of the Optimatch UD search tool from the German registry (ZKRD) including the possibility to select for HLA-DPB1 permissive donors |
| 70 | N/A | N/A | 2016 | N/A | Description of the HapLogic UD search tool from the US registry (NMDP) including the possibility to select for HLA-DPB1 permissive donors |
| 71 | 416 | AL, MDS | 2005-2014 | 10/10; HLA-DPB1 nonpermissive TCE or ΔFD between patient and donor; ΔFD > 2.665: worse OS and EFS due to less relapse and less NRM; better predictive than nonpermissive TCE in this cohort | Refinement of nonpermissive HLA-DPB1 TCE mismatches by ΔFD scores might further improve the predictive value of the algorithm |
| Myeloablative conditioning | |||||
| T-cell depletion and T-cell replete UD-HCT | |||||
| 75 | N/A | AML | N/A | Development of an in vitro protocol to generate CD4+ T cells alloreactive to mismatched HLA-DPB1 on AML blasts or from RNA-transfected dendritic cells; preclinical humanized mouse model of GVL targeted to HLA-DPB1 | First protocol for the isolation and expansion of CD4+ T cells from the naive repertoire using primary AML blasts or transfected autologous dendritic cells for cellular immunotherapy; first demonstration of the in vivo efficacy of CD4+ T cells alloreactive to HLA-DPB1 in mediating GVL against AML |
| 67 | 595 | Patients searching for an UD | N/A | 10/10; probability to find an HLA-DPB1 allele-matched or permissive UD | 70% of patients have a 10/10, young, HLA-DPB1 TCE permissive UD |
| Reference . | No. . | Patient and HCT characteristics . | Era . | HLA matching* and clinical outcomes . | Major findings/impact . |
|---|---|---|---|---|---|
| 46 | 1 | CML | 2001 | 11/12 in GVH direction; case report, acute rejection | First evidence that mismatched HLA-DPB1 can be the target of CD4+ T-cell–mediated allograft rejection |
| Case report | |||||
| 47 | 143 | Various diseases | 1996-2001 | 10/10; HLA-DPB1 allele mismatches; increased aGVHD, decreased relapse | First evidence of a GVL effect targeted to HLA-DPB1 allele mismatches after UD-HCT |
| Myeloablative conditioning | |||||
| T-cell depletion | |||||
| 62 | 118 | Various diseases | 1995-2002 | 10/10; HLA-DPB1 nonpermissive TCE mismatches; increased aGVHD and NRM | First description of a T-cell epitope matching score for HLA-DPB1 |
| Myeloablative conditioning | |||||
| T-cell depletion | |||||
| 50 | 423 | Various diseases | 1996-2003 | 10/10; HLA-DPB1 allele mismatches; increased aGVHD, decreased relapse, better OS in ALL | Confirmation of Shaw et al47 and possibly improved survival of ALL patients after HLA-DPB1 mismatched HCT |
| Myeloablative conditioning | |||||
| T-cell depletion | |||||
| 55 | 72 | Thalassemia | 1992-2004 | 10/10; HLA-DPB1 nonpermissive HVG mismatches; increased graft rejection | First association of nonpermissive TCE mismatches with outcome of HCT for nonmalignant disease |
| 79 | N/A | N/A | N/A | N/A | First protocol of targeted epitope-specific HLA-DPB1 typing for UD-HCT |
| 38 | 5929 | Various diseases | 1984-2005 | 5-8/8; HLA-DPB1 allele mismatches; increased aGVHD, decreased relapse | First definitive confirmation of GVL targeted to HLA-DPB1 allele mismatches after UD-HCT in a T-cell–replete cohort |
| Registry study (14th IHIW) | |||||
| 53 | 1 | Lymphoblastic lymphoma | 2006 | 10/10; 1 permissive and 1 nonpermissive HlA-DPB1 TCE mismatch | First isolation of CD4+ T cells alloreactive to mismatched HLA-DPB1 from a patient clearing residual malignant disease after donor lymphocyte infusion from an UD |
| Case report | |||||
| 63 | 621 | Various diseases | 1999-2006 | 10/10; HLA-DPB1 nonpermissive TCE mismatches; increased aGVHD and NRM, worse OS | First demonstration of a negative impact of nonpermissive HLA-DPB1 mismatches in a large multicenter registry cohort |
| Myeloablative and reduced-intensity conditioning | |||||
| Registry study (IBMDR) | |||||
| 51 | 488 | AML, ALL, CML | 1996-2006 | 10/10 and <10/10; HLA-DPB1 allele mismatches; decreased relapse, increased NRM, worse OS in early disease in 10/10; improved OS in late disease in <10/10 | Differential impact of HLA-DPB1 allele mismatches in 10/10 or <10/10 according to disease status |
| Myeloablative and reduced-intensity conditioning | |||||
| T-cell depletion | |||||
| 49 | 4 | Healthy donors | N/A | In vitro generation of 16 T-cell clones alloreactive to a variety of HLA-DP specificities by stimulation with transfected HELA cells | Confirmation that CD4+ T-cell alloreactivity to HLA-DPB1 can be elicited by both permissive and nonpermissive mismatch combinations |
| 65 | 24 | Healthy donors | N/A | In vitro mixed lymphocyte reactions between 10/10 matched, HLA-DPB1 permissive or nonpermissive UD | First experimental evidence for more potent T-cell alloreactivity to nonpermissive than to permissive HLA-DPB1 TCE mismatches |
| 56 | 115 | AML, ALL, CML, MDS, UD-HCT | 1990-2002 | 6/12 to 12/12; retrospective matched control study graft failure vs engraftment | Graft failure is associated with donor-specific HLA antibodies, over half of them against HLA-DP |
| 57 | 592 | AML, ALL, MDS and others, UD-HCT | 2005-2009 | 9/12 to 12/12; prospective HLA antibody testing | Donor-specific antibodies in 1.4% of patients, all against mismatched HLA-DP; significantly associated with graft failure |
| 39 | 8539 | AML, ALL, CML, MDS | 1993-2007 | 10/10 and 9/10; HLA-DPB1 nonpermissive TCE mismatches; increased aGVHD and NRM, worse OS | First definitive demonstration of a negative impact of nonpermissive HLA-DPB1 mismatches on OS in 10/10 and 9/10 UD-HCT |
| Registry study (15th IHIW) | |||||
| 54 | 24 | Various diseases | 2000-2008 | 10/10; CD4+ DLI for mixed chimerism or relapse; ex vivo analysis of T cells alloreactive to HLA-DPB1 in responding or nonresponding patients | First demonstration of reliable emergence of CD4+ T cells alloreactive to HLA-DP in patients responding to DLI after UD-HCT |
| T-cell–depleted UD-HCT | |||||
| 68 | N/A | N/A | N/A | N/A | Launch of the first free online tool for HLA-DPB1 nonpermissive/permissive TCE match assignment |
| 36 | 2 | AML | N/A | 10/10; HLA-DPB1 nonpermissive GVH mismatch in both patients; severe aGVHD of the gut after CMV reactivation | Host-derived CMV-specific T cells can trigger CD4+ T-cell alloreactivity against mismatched HLA-DPB1 after UD-HCT, leading to severe GVHD |
| 2 case reports | |||||
| 66 | 1281 | AML, ALL, CML, MDS | 1988-2003 | 10/10; HLA-DPB1 nonpermissive GVH mismatches significantly lower relapse than permissive mismatches; no impact of concomitant mismatching for DPA1 | GVL effect by nonpermissive GVH mismatches; no impact of DPA1 on nonpermissive TCE mismatches |
| Registry study (CIBMTR) | |||||
| 4 | 8003 | AML, ALL, CML, MDS | 1999-2011 | 10/10 and 9/10; HLA-DPB1 nonpermissive TCE mismatches; increased NRM, worse OS in 10/10 but not in 9/10 | First confirmation of a negative impact of nonpermissive HLA-DPB1 mismatches in 10/10 on OS in an independent validation cohort from a registry |
| Registry study (CIBMTR) | |||||
| 71 | 4 | Healthy donors | N/A | Generation of 12 site-directed mutants of HLA-DPB1*09:01 at 10 key amino acid positions; investigation of their allorecognition by 17 T-cell effectors alloreactive to wild-type HLA-DPB1*09:01; determination of the median impact of each substitution on T-cell alloreactivity (FD) | Development of a FD score system for individual amino acid polymorphism in HLA-DPB1, and for HLA-DPB1 alleles. The FD score of any current or future HLA-DPB1 allele can be used to predict its assignment to TCE groups |
| 52 | 7898 | Various diseases | 1993-2010 | 10/10; role of HLA-locus mismatches including HLA-DPB1 allelic disparity; increased aGVHD, decreased relapse | Confirmation of lower relapse risk associated with HLA-DPB1 allele mismatches; no such association was observed with HLA-DRB1 allele mismatches |
| Registry study (JMDP) | |||||
| 64 | 2029 | AML, ALL, CML, MDS | 1988-2008 | 10/10; truly unidirectional GVH HLA-DPB1 expression mismatches; increased aGVHD when patient carries high-expression mismatch | First demonstration that expression levels determine nonpermissive GVH HLA-DPB1 mismatches; conclusive association between 3′ UTR expression SNPs and exon HLA-DPB1 types |
| Registry Study (CIBMTR) | |||||
| 80 | 1342 | Various diseases | 2000-2008 | 10/10; increased grade III-IV aGVHD with 2 allelic HLA-DPB1 mismatches, increased relapse with TCE nonpermissive HVG disparities | HLA-DPB1 impacts aGVHD and relapse by number of allelic mismatches and TCE HVG disparities, respectively |
| Registry study (RFGM) | |||||
| 69 | N/A | N/A | 2013 | N/A | Description of the Optimatch UD search tool from the German registry (ZKRD) including the possibility to select for HLA-DPB1 permissive donors |
| 70 | N/A | N/A | 2016 | N/A | Description of the HapLogic UD search tool from the US registry (NMDP) including the possibility to select for HLA-DPB1 permissive donors |
| 71 | 416 | AL, MDS | 2005-2014 | 10/10; HLA-DPB1 nonpermissive TCE or ΔFD between patient and donor; ΔFD > 2.665: worse OS and EFS due to less relapse and less NRM; better predictive than nonpermissive TCE in this cohort | Refinement of nonpermissive HLA-DPB1 TCE mismatches by ΔFD scores might further improve the predictive value of the algorithm |
| Myeloablative conditioning | |||||
| T-cell depletion and T-cell replete UD-HCT | |||||
| 75 | N/A | AML | N/A | Development of an in vitro protocol to generate CD4+ T cells alloreactive to mismatched HLA-DPB1 on AML blasts or from RNA-transfected dendritic cells; preclinical humanized mouse model of GVL targeted to HLA-DPB1 | First protocol for the isolation and expansion of CD4+ T cells from the naive repertoire using primary AML blasts or transfected autologous dendritic cells for cellular immunotherapy; first demonstration of the in vivo efficacy of CD4+ T cells alloreactive to HLA-DPB1 in mediating GVL against AML |
| 67 | 595 | Patients searching for an UD | N/A | 10/10; probability to find an HLA-DPB1 allele-matched or permissive UD | 70% of patients have a 10/10, young, HLA-DPB1 TCE permissive UD |
aGVHD, acute GVHD; AL, acute leukemia; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CIBMTR, Center for International Blood and Marrow Transplant Research; CML, chronic myeloid leukemia; CMV, cytomegalovirus; DLI, donor lymphocyte infusion; EFS, event-free survival; IBMDR, Italian Bone Marrow Donor Registry; IHIW, International Histocompatibility Workshop; JMDP, Japanese Marrow Donor Program; MDS, myelodysplastic syndrome; N/A, not applicable; NMDP, National Marrow Donor Program; NRM, nonrelapse mortality; OS, overall survival; RFGM, Registre France Greffe de Moelle; UTR, untranslated region; ZKRD, Zentrales Knochenmarkregister Deutschland.
HLA matching is reported as the number of matched HLA-A, -B, -C, -DRB1, -DQB1 alleles (10 of 10 or other), followed by the algorithm used to consider HLA-DPB1 mismatches (alleles, nonpermissive TCE, or other).
HLA-DPB1 as a model for permissive mismatches
The concept of permissive HLA mismatches in allo-HCT is the subject of intense debate, and has sometimes been erroneously interpreted as mismatches not eliciting any T-cell response which, due to the nature of alloreactivity, is unlikely. Instead, the notion of permissive mismatches is based on accumulating evidence suggesting that limited alloreactivity is sufficient for GVL, whereas aggressive alloreactivity can lead to clinically uncontrollable GVHD.59,60 Permissive mismatches are those eliciting limited alloreactivity, shifting the balance from GVHD to GVL with good clinical tolerability while maintaining treatment efficacy.61
HLA-DPB1 is particularly amenable to investigating permissive mismatches in UD-HCT, due to the shuffling of tightly linked hypervariable SNP regions both inside and outside of the exons (as mentioned in “Structure and polymorphism of HLA-DP”). Two different, though partly overlapping, models have been proposed to this regard (Table 1). The first is based on structural T-cell epitopes (TCEs) shared by subsets of HLA-DPB1 alleles (TCE model)39,62,63 ; the second considers differences in HLA-DPB1 allele cell-surface expression levels (expression model).64 The TCE model stems from experimental data showing that T cells alloreactive to HLA-DPB1*09:01 reproducibly cross-recognize other nonself HLA-DPB1 alleles from at least 3 distinct TCE groups. It was predicted that mismatches between HLA-DPB1 alleles from the same TCE group (designated permissive) would elicit less vigorous T-cell alloreactivity compared with alleles from different TCE groups (designated nonpermissive). This hypothesis was proven correct, both experimentally65 and clinically, with nonpermissive HLA-DPB1 mismatches in either GVH or HVG direction resulting in worse survival after HLA-matched UD-HCT.4,39,62,63 The survival disadvantage of nonpermissive mismatches is the net result of a greater risk of severe acute GVHD without a significant change in relapse risk compared with permissive mismatches.39,66 It should be noted that alterations in GVL do not apply to transplants for nonmalignant disorders and these are therefore best performed using an HLA-DPB1 allele-matched UD.55 In contrast, current guidelines for patients transplanted for malignant disease recommend selection of an HLA-DPB1 allele-matched or permissively mismatched UD, available with a probability of at least 70%.67 Thus, by applying the concept of permissive TCE matching, the chances of finding an “intelligently” mismatched UD has significantly increased, and online bioinformatics tools have been made available by donor registries in the United States and Europe to facilitate TCE selection in clinical practice.68-70 Concomitantly, requests for HLA-DPB1 typing upfront in clinical UD searches through the NMDP have greatly increased from 11% to 57% over the last decade (Jason Dehn, NMDP, personal communication, 6 March 2017). Recently, it was shown that the in silico assignment of HLA-DPB1 alleles to TCE groups can be performed based on their overall “functional distance” (FD) score, that is, the median impact of 12 key amino acid polymorphisms on T-cell alloreactivity.71 A further refinement of the TCE model was subsequently proposed by the novel concept of ΔFD, that is, the net difference between FD scores of mismatched HLA-DPB1 alleles in patient and donor, which has an 80% overlap with the original TCE model and awaits validation in further independent studies.72 The expression model is based on the observation that the 2 variants of a biallelic SNP in the HLA-DPB1 3′ untranslated region are associated with high or low HLA-DP expression levels, respectively,73 and that these variants in turn are in tight LD with specific HLA-DPB1 alleles. The expression model was found to be predictive of GVHD in the particular situation of truly single HLA-DPB1 mismatches, that is, 1 HLA-DPB1 allele shared between patient and donor and the other 1 mismatched at least in the GVH vector, in which the latter is high expression in the patient but low expression in the donor.64 The deleterious situation in the expression model is therefore present only in a subset of transplants, whereas both GVH and HVG mismatches across different TCE groups are deleterious in the TCE model. Interestingly, SNP-based assignment of the most frequent high- or low-expression HLA-DBP1 alleles closely parallels their TCE assignment, raising the intriguing possibility that the 2 models might be surrogates or synergistic.74 Further work is warranted to fully characterize this relationship.
New opportunities for cellular immunotherapy targeted to mismatched HLA-DP
The characteristics of mismatched HLA-DP to mediate GVL by alloreactive CD4+ T cells open new horizons for their exploitation in adoptive cellular immunotherapy of malignant blood disorders. To this end, a protocol for isolation of cytolytic CD4+ T cells alloreactive to a broad range of HLA-DP specificities from the CD45RA-naive pool, able to eradicate acute myeloid leukemia in a preclinical murine model, has recently been described.75 T cells with direct alloreactivity can also be found in the memory pool,9,32,76-78 which might be an even better reservoir given the potential of memory T cells to concomitantly improve immune reconstitution against infective agents. Our extensive understanding of permissive mismatches will help us to carefully select optimal donor-recipient HLA-DPB1 mismatch combinations to minimize the risk of GVHD while maintaining efficient GVL in protocols of adoptive cellular immunotherapy, which hold promise to provide further advances in curing malignant hematological diseases.
Conclusion
HLA-DP disparity is both a blessing and a bane in HLA-A, -B, -C, -DRB1-identical UD-HCT and represents a fundamental biological difference from sibling HCT where it is generally absent. Increased knowledge of the mechanisms underlying permissive HLA-DP mismatches which mediate limited T-cell alloreactivity and thereby shift the balance from GVHD toward GVL, will pave the way to their exploitation in cellular immunotherapy of malignant hematological disorders.
Acknowledgments
The authors thank Jason Dehn for the NMDP HLA-DP typing percentages.
This work was supported by grants from the Deutsche José Carreras Leukämie Stiftung (DJCLS R 15/02), the European Commission Transcan JTC2012 (Cancer12-045-HLALOSS), the Dr Werner Jackstädt Stiftung, and the Joseph Senker Stiftung (K.F.).
Authorship
Contribution: K.F. and B.E.S. wrote the manuscript and created the figure and table.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katharina Fleischhauer, Institute for Experimental Cellular Therapy, Essen University Hospital, Institutsgruppe 1 (IG1), 11th Floor, Hufelandstraße 55, D-45122 Essen, Germany; e-mail: katharina.fleischhauer@uk-essen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal