Abstract
The heterogeneous nature of acute myeloid leukemia (AML) and its poor prognosis necessitate therapeutic improvement. Current advances in AML research yield important insights regarding AML genetic, epigenetic, evolutional, and clinical diversity, all in which dysfunctional p53 plays a key role. As p53 is central to hematopoietic stem cell functions, its aberrations affect AML evolution, biology, and therapy response and usually predict poor prognosis. While in human solid tumors TP53 is mutated in more than half of cases, TP53 mutations occur in less than one tenth of de novo AML cases. Nevertheless, wild-type (wt) p53 dysfunction due to nonmutational p53 abnormalities appears to be rather frequent in various AML entities, bearing, presumably, a greater impact than is currently appreciated. Hereby, we advocate assessment of adult AML with respect to coexisting p53 alterations. Accordingly, we focus not only on the effects of mutant p53 oncogenic gain of function but also on the mechanisms underlying nonmutational wtp53 inactivation, which might be of therapeutic relevance. Patient-specific TP53 genotyping with functional evaluation of p53 protein may contribute significantly to the precise assessment of p53 status in AML, thus leading to the tailoring of a rationalized and precision p53-based therapy. The resolution of the mechanisms underlying p53 dysfunction will better address the p53-targeted therapies that are currently considered for AML. Additionally, a suggested novel algorithm for p53-based diagnostic workup in AML is presented, aiming at facilitating the p53-based therapeutic choices.
Introduction
Acute myeloid leukemia (AML) is a genetically heterogeneous clonal hematopoietic stem cell malignancy characterized by chromosomal abnormalities, recurrently mutated genes, epigenetic modifications affecting chromatin structure, and microRNAs deregulations. Despite progress in understanding AML pathobiology, the still-existing poor outcome, especially in the elderly, obligates therapeutic improvement. Major obstacles impacting treatment efficacy result from AML genomic heterogeneity, interpatient variability, and the apparent need to address various AML targets.1,2 In this context, p53 and its related network deserve special consideration.
p53 plays a pivotal role in normal and leukemic hematopoiesis and is central in a complex web of AML-related signaling pathways. AML genetic and epigenetic aberrations encompass the p53 gene, protein, and regulated network, among others.
The causal relation between leukemogenesis and p53 dysfunction has been demonstrated in Li-Fraumeni syndrome (LFS), large-scale sequencing studies, proteomics, and clinical and laboratory settings.3-10 Currently, TP53 mutations, per se, are prioritized over other p53 abnormalities with regard to AML pathogenesis, therapy response, and prognosis. However, evolvement in the field of p53 research has modified our view of p53 dysfunction by elucidating the mutant p53 (mutp53) gain-of-function (GOF) phenomenon11 and by unveiling molecular mechanisms underlying nonmutational wtp53 inactivation.
Because TP53 mutations occur in only ∼8% of de novo AML cases and LFS-related AML is conspicuously rare, other nonmutational p53 abnormalities might bear a greater AML-related significance than is currently appreciated.3,4
Indeed, recent data indicate that nonmutational wtp53 dysfunction may occur in nearly all AML subsets (European Leukemia Net classification). The inactivating mechanisms reach beyond the 2 already well recognized, namely MDM2/4 overexpression and p14ARF inactivation, and might bear potential therapeutic relevance.
Accordingly, we advocate herein the need to evaluate p53 genotype and functional phenotype during AML workup, thus adding an extra platform for precision diagnosis and treatment in AML.
Elucidating the unique pattern of mutational or nonmutational p53 dysfunction in the individual AML patient may promote rationalizing the p53-based therapeutic approach consisting of mutp53 reactivation, abrogation of mutp53 GOF underlying mechanisms, or rescuing wtp53 inactivation.12,13
For advancing this goal, we discuss AML-associated TP53 mutations and mutational and nonmutational p53 function-disrupting mechanisms (Figure 1). We map them to the various AML entities and revise therapeutic options targeted to address them. Additionally, a diagnostic algorithm for p53 profiling, aiming to support personalized p53-based therapeutic decisions, is proposed.
Role of p53 in hematopoiesis
The tumor suppressor p53 transcription factor plays a key role in preserving genomic integrity, thereby preventing tumorigenesis.
p53 also regulates additional cellular processes, including maintenance of normal stem cell pool, serving as a barrier to cancer stem cell formation.14 In hematopoietic stem/progenitor cells (HSPCs), p53 is actively involved in regulating their quiescence and self-renewal, thus averting leukemogenesis.8 The involved mechanisms include p53’s negative interaction with telomerase and its negative impact on microenvironment-mediated LSCs survival.15,16
Hence, p53 inactivation promotes HSPCs proliferation, leading to DNA damage accumulation and the malignant transformation of HSPCs.8
TP53 mutations in AML
While AML genomes are conspicuously characterized by fewer mutations than most other adult cancers, mutations in tumor suppressor genes, including TP53, comprise 16% of the key functional categories of de novo AML genetic aberrations.4,17
TP53 mutations are early leukemogenic initiating driver mutations, as demonstrated in clonal hematopoiesis, myelodysplastic syndrome, complex karyotype (CK) AML, and therapy-related AML (t-AML), wherein TP53 mutations are detected in the founding clones.5,7,18-20
The clinical significance of TP53 mutations in nonleukemic “rising clones” following induction therapy and of those emerging while on MDM2 inhibitors (MDM2i) therapy in relapsed/refractory AML remains to be determined.21,22 Anecdotal cases of late appearance of TP53 mutation in wtp53 AML patients during follow-ups might indicate emergence of t-AML.23
The early location of TP53 mutation in the leukemic clonal hierarchy is of major relevance for disease pathogenesis and therapeutic response. AML with TP53 mutations in subclones only is thus expected to display differential susceptibility to p53-targeted therapies.24
AML-related TP53 mutations and/or copy-number aneuploidies (CNAs) were recently categorized as a distinct driver lesion, reflecting discrete paths in AML evolution, and defining a separate molecular AML subgroup with adverse prognosis.5 These mutations underlay an aggressive disease, chemoresistance, and dismal outcome even after allogeneic HSPC transplantation.25,26
AML-related TP53 mutations differ in their frequency, characteristics, and functional impact. Notably, TP53 mutational allele burden (or variant allele frequency [VAF]) has been found to predict outcome in myelodysplastic syndrome and secondary AML. This finding, though unconfirmed, requires further evaluation addressing the prognostic value of mutational burden levels and their dynamic course.27,28
TP53 mutations occur in only 8% of de novo AML cases, presenting one of the lowest frequency rates in comparison with other malignancies.4,17 Contrarily, in t-AML, wherein TP53 is the most commonly mutated gene, TP53 mutation frequency is ∼30%. Interestingly, for decades, the increased frequency of TP53 mutations in t-AML was attributed to their induction by chemotherapy. Recently, however, it has been indicated that cytotoxic therapy does not directly induce TP53 mutations. Rather, it confers survival advantage to rare (0.003% to 0.7%) chemotherapy-resistant HSPCs (in a founding clone) carrying age-related TP53 mutations that expand preferentially after chemotherapy.20
In CK-AML, wherein TP53 mutations are the most common molecular lesion, the TP53 mutation rate is ∼70%.26 The increased CK frequency in elderly de novo AML patients, AML postmyeloproliferative disorders, and t-AML correlates with the increased TP53 mutation rate in these entities (21% to 33%), hence denoting mutp53-induced genomic destabilization.20,25,29,30 Another determinant of the rate of TP53 mutations in AML is their mutual exclusivity with other mutated genes (NPM1, FLT3, MDM2, and ARF).4,31 Conversely, past chemotherapy, chemoresistance, and cancer family history imply the possible presence of TP53 mutations. Notably, only 1% to 3% of LFS-related tumors carrying TP53 germ line mutations are leukemias (mostly acute lymphoblastic leukemia, rarely AML [mainly t-AML]). Studies addressing this infrequency focus on environmental and genetic modifiers that might promote LFS-related AML, including chemotherapy, ionizing radiation, single-nucleotide polymorphisms (SNPs) in TP53 pathway genes, increased CNAs, and telomere attrition.3 Possible underdiagnosed LFS in t-AML should be ruled out by prospective evaluation of TP53 germ line mutations or deletion in t-AML cases.32
AML-related TP53 mutations are mostly missense somatic mutations that embrace some of the most frequent cancer-associated TP53 hotspots, such as the contact mutations at codons 248, 273, and the structural mutation at codon 245.33
Next-generation sequencing (NGS) studies indicate that mutp53-AML cells display mostly heterozygous mutations.34 However, the heterozygous status is often unstable, as TP53 mutations are frequently followed by loss of heterozygosity (LOH) during cancer progression. Indeed, p53 LOH is frequently found in t-AML, AML postmyeloproliferative Ph1-negative neoplasms, and in LFS-related AML.3,30,35 The most frequent TP53 mutation signature in AML is a G:C >A:T transition mutation, occurring at the hypermutable CpG dinucleotides with a frequency of 23.3%.33 This age-correlated signature (1A/B) ensues presumably, an endogenous mutational process.36 Notably, except for the aforementioned mutual exclusivity, no unique TP53 mutation–associated gene signatures have been discovered yet.7,11
Besides TP53 mutations, few SNPs in the TP53 locus, such as P47S and R72P, exhibit cancer-related phenotypical manifestations. Currently, it is well accepted that TP53-R72 is more effective at inducing cell death and protecting cells from malignant transformation than the TP53-P72 variant. Additionally, SNPs in TP53 pathway genes (eg, MDM2) might also have biological consequences, either individually or in combination with TP53.37 Indeed, a synergistic effect of the TP53-P72 and MDM2 SNP309-G alleles, which increase t-AML risk, has been found.38,39
Mechanisms underlying mutp53 LOF/GOF in AML
TP53 mutations modify wtp53 protein structure, folding, and stability, thus abrogating its DNA binding and activity.
Loss of function (LOF) is frequent among p53 missense mutants.11 Indeed, germ line TP53 LOF mutations, underlying LFS, abrogate TP53 function as a gatekeeper gene, thus predisposing to a variety of early-onset malignancies.3
Likewise, somatic TP53 LOF mutations promote HPSC clonal expansion and drive AML initiation.18,20,25,40
The frequency of mutp53 GOF, which empowers mutp53 tumorigenicity and promotes unfavorable AML biology, has not been assessed yet. Nevertheless, different p53 mutants acquire GOF, which vary in their effect and magnitude as a function of mutation site and nucleotide substitution type.41 Hence, mutp53 represents a heterogeneous protein population with divergent potential to serve as “druggable” targets (Figure 2).11 The major cellular impacts of mutp53 GOF in AML include genomic destabilization and disruption of the DNA damage response (DDR), loss of cell-cycle control, enhanced tumor cell proliferation, and chemoresistance.
Unlike wtp53, which exerts its tumor-suppressive activity primarily via transactivation of downstream target genes, most GOF properties stem from binding of mutp53 to various cellular proteins, including transcription factors.11,42
Mutp53 binding to HSP90 (overexpressed in poor prognosis AML) inactivates MDM2 and CHIP E3 ligase–mediated p53 degradation.
The resultant mutp53 stabilization and accumulation in cancer cells is a key requisite for GOF acquisition.43-45 Another key protein–protein interaction is the TopBP1-mediated mutp53-p73 binding, which abrogates p73 function, blocks its DNA binding activities, inactivates p53-independent apoptosis, and causes inefficient spindle checkpoint function and aneuploidy.11,42,46 Additionally, several mutp53 interact with Mre11-Rad50-NBS1 complex thus interfering with DNA-damage signaling, and with ETS1, thus activating multidrug resistance 1 (MDR1) transcription.42,47 The resultant P-gP upregulation confers multidrug resistance (MDR) to various AML medications.47,48 Indeed, activated MDR1 is recognized as an independent prognostic variable for induction failure in adult AML.48
Mutp53 also augments TNFα-induced NF-κB activation and influences NF-κB target gene transcription. Aberrant NF-κB activation reduces apoptosis, favors cell proliferation, and enhances MDR1 expression. Hence, copresence with p53 GOF mutants intensifies dysregulation of these cell cycle events and promotes chemoresistance.41,49-51
Mutp53 GOF promotes genomic instability and defective DDR
Mutp53 GOF promotes chromosomal instability comprising aneuploidy and structural changes that facilitate LOH and amplification instability.46 Indeed, mutp53-AML is frequently associated with chromosomal aneuploidy (mainly −5/5q, −7/7q, and −17/17p) and with CK, wherein chromothripsis occurs in ∼50% of cases.5,26,34,52 The genomic instability is mediated mostly through mutp53 GOF-induced modifications in DDR and elevated ROS.
The underlying mechanisms include the aforementioned interaction with p73 and with DNA topoisomerase I/cyclin A1, MRE11, centrosome proteins, and telomere capping proteins. The Mutp53–DNA topoisomerase I interaction, mediated by cyclin A1 (overexpressed in AML) via site-specific mutp53 phosphorylation, yields a stable complex. The resultant constitutive topoisomerase I stimulation and the consequent aberrant homologous DNA recombination lead to genomic instability.53 Additionally, AML-related p53 GOF mutants interact physically with MRE11 and disrupt recruitment of the MRE11– RAD50–NBS1 complex to DNA double-stranded breaks, thus inactivating MRE11/ATM-dependent DDR.42 Moreover, p53 GOF mutants attenuate antioxidant enzymes, thereby contributing to AML-related ROS accumulation while promoting cell survival and abrogating the DDR.41,54
Mutp53 GOF promotes cell proliferation and chemoresistance
Loss of cell-cycle control and tumor cell proliferation are facilitated by the interaction between p53 GOF mutants and p73, NF-κB, PML, and ETS2, which activates the MLL1/2 and MOZ chromatin regulatory genes.55 Notably, the MLL1/MOZ-related AML entities, namely AML with t(11q23), inv(8), and t(8;16), are accompanied by dysfunctional wtp53 rather than mutp53, hence indicating the possible link between abrogated p53 and AML-related chromatin changes.
Chemoresistance is advanced by the interaction between p53 GOF mutants and ETS1 and NF-κB, which upregulate MDR1 transcription, and by their antiapoptotic function. The intrinsic apoptotic pathway is abrogated by mutp53-mediated downregulation of proapoptotic genes (Bax, Puma, and Noxa) via mutp53–p73/63 complexing, which blocks p73/p63 DNA binding, and by mutp53-induced upregulation of antiapoptotic genes (BCL-XL and EGR1).11,42
The extrinsic pathway is affected by mutp53-induced repression of the proapoptotic CD95 (Fas/Apo-1), yielding partial alleviation of CD95-dependent cell death.11 Importantly, although CD95 is expressed in AML with variable intensity, the predictive value of its expression for chemotherapy-induced remission is controversial.56 Notably, p53-independent apoptosis, stimulated by specific chemotherapeutics (eg, doxorubicin),57 might also be abrogated by the mutp53–p73 interaction.
Genomic loss of TP53 in AML
The critical role of p53 dysfunction in AML is highlighted also by loss of TP53 locus, which promotes AML evolution. In vitro studies have shown that wtTP53 introduction into the p53-nonproducer promyelocytic leukemia cell line HL-60 induces promyelocyte differentiation into granulocytic/monocytic lineages and increases chemotherapy sensitivity.9,58 The complexity of TP53 loss–related leukemogenesis was demonstrated in genetically engineered mice model in which a concomitant haploinsufficiency of Egr1 and APC was required for induction of AML with low penetrance and long latency.10
Deletion of 17p, to which TP53 is mapped, occurs in AML as a single aberration or with additional chromosomal abnormalities, indicating an unfavorable cytogenetic category.2,59 The detrimental effect of 17p− is implicated to reflect a joined impact of TP53 loss and the reduced dosage of linked tumor suppressor genes.60 Notably, biallelic loss of TP53 is a rare event in AML that might be addressed by gene transfer therapy, which is currently under preclinical evaluation.13
Nonmutational wtp53 inactivation
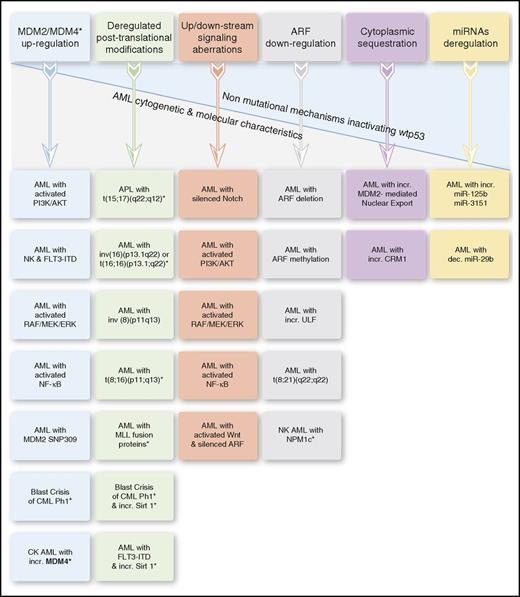
Lack of normal p53 activities affects AML biology and treatment also in the presence of nonmutational wtp53 inactivation. This might occur in various AML entities via diverse inactivating mechanisms, affecting p53 protein levels, posttranslational modifications, subcellular localization, and signaling pathways and regulators, including the p53-microRNAs (miRs) (Figure 3).
Nonmutational mechanisms inactivating wtp53 in AML. The asterisk in the “Deregulated posttranslational modifications” column denotes deacetylated p53. In certain AML entities, wtp53 inactivation results from ≥1 underlying mechanism. APL, acute promyelocytic leukemia; dec., decreased; incr., increased; NK, normal karyotype.
Nonmutational mechanisms inactivating wtp53 in AML. The asterisk in the “Deregulated posttranslational modifications” column denotes deacetylated p53. In certain AML entities, wtp53 inactivation results from ≥1 underlying mechanism. APL, acute promyelocytic leukemia; dec., decreased; incr., increased; NK, normal karyotype.
The frequency, mechanism, and oncogenic power of nonmutational wtp53 inactivation in AML still need to be evaluated.
While mutp53-AML commonly co-occurs with −5/5q−, −7/7q−, −17/17p−, these cytogenetic abnormalities do not co-occur in AML entities with possible p53 dysfunction, such as mutNPM1, PML-RARA, t(8;21), and inv(16) leukemias (hereinafter). Also, PML-RARA and mutNPM1 commonly co-occur with FLT3 mutations, which are very unusual for mutp53-AML. This suggests that p53 dysfunction in these AML entities contributes to leukemogenesis in different ways than TP53 mutations. Nevertheless, the adverse cytogenetics and poor outcome, which do occur in wtp53-AML with MDM2/4 or CRM1 overexpression, suggest that nonmutational wtp53 dysfunction might indeed have significant oncogenic potential.
AML with deregulated ARF-MDM2/4-p53 axis
The ARF-MDM2/4-p53 axis is abrogated in most AML cases, undermining wtp53 function due to ARF inactivation and/or MDM2/4 overexpression. Normally, ARF, a major wtp53 positive regulator, antagonizes its MDM2-dependent degradation. While ARF deletion is uncommon in AML (5%), low ARF expression occurs frequently (>40%) due to increased ULF messenger RNA (mRNA) levels, aberrant methylation of ARF promoter (rare), and attenuating effects of AML1-ETO and NPM1c+ proteins.61-64 Importantly, attenuated ARF contributes also to wtp53 dysfunction associated with other aberrantly activated signaling pathways detailed below.
MDM2 overexpression is frequently observed in AML (up to 50%), ensuing from activated phosphatidylinositol 3-kinase (PI3K)/AKT, RAS/RAF/MEK/extracellular signal-related kinase (ERK), and NF-κB pathways; MDM2 SNP309 polymorphism; and increased MDM2 mRNA translation. It is a major wtp53 inactivating mechanism, as it inhibits wtp53 transcriptional activity, mediates its degradation, and regulates its subcellular localization.6,61,65-68
MDM4, another wtp53 negative regulator, is overexpressed in 10% of wtp53 CK-AML. In the OCI/AML-2 cell line, it suppresses p53 activity, decreases its half-life, and promotes its cytoplasmic sequestration.69,70 Notably, MDM2/4 overexpression in AML represents a molecular setup in which dysfunctional wtp53 is associated with unfavorable cytogenetics and poor prognosis, indicating the potential deleterious effect of nonmutational wtp53 dysfunction on genomic stability.6,61,65
AML with NPM1 and FLT3 mutations
NPM1/FLT3-mutated AML is often accompanied by wtp53 dysfunction due to several inactivating processes.
Cytoplasmic mutNPM1 (NPM1c+) inactivates wtp53 by abrogating the ARF-MDM2-p53 axis. It complexes with wtNPM1 and with ARF leading to their cytoplasmic mislocalization and ULF-mediated ARF ubiquitination. The resultant attenuation in ARF-induced p53 stabilization promotes its MDM2-mediated degradation.71 Interestingly, mutNPM1-AML is also characterized by miR-10a overexpression, which is associated with MDM4 downregulation and highly expressed p53β/γ isoforms, which most likely contribute to the relative good prognosis of this AML entity.72,73
FLT3-ITD mutations, predicting poor AML outcome, are associated with wtp53 dysfunction via SIRT1 overexpression, which yields p53 deacetylation,74 PI3K/AKT pathway activation (which promotes MDM2-mediated p53 degradation),66 and STAT/MAPK pathway activation with Bcl2 accumulation, which opposes p53 activity.75
AML with chromosomal translocations
Fusion proteins (FPs) ensuing chromosomal translocations abrogate wtp53 activity via numerous mechanisms, among which wtp53 deacetylation is the most frequent.
The PML-RARα FP of t(15;17) APL disrupts the activities of wtPML, an upstream p53 regulator, and mediates histone deacetylase (HDAC) recruitment, thus triggering p53 deacetylation and MDM2-mediated degradation.76 Indeed, APL mouse models have demonstrated that ATRA and arsenic-trioxide eradicate APL by inducing PML-RARα degradation and PML-p53 axis activation.77
p53 deacetylation is also triggered by the CBFβ-SMMHC FP of inv(16)/t(16;16) AML, which complexes with HDAC8 and p53, thus promoting p53 deacetylation.78 Additionally, the CBFβ-SMMHC/Runx1 oligomers mislocalize HIPK2 (p53 activator) into cytoplasmic filamentous structures, hence interfering with p53 apoptotic function.79 The favorable prognosis of this AML and of APL might be explained in part by the presence of a relatively higher TAp73/ΔNp73 ratio, compensating for wtp53 dysfunction.80
Conversely, t(11q23) AML, generating several FPs (MLL-AF9, MLL-AF10, MLL-ENL, and MLL-ELL) that bind to p53 and interfere with its p300-mediated acetylation, is of poor prognosis.81
The MOZ-CBP (CREB-binding protein) FP of t(8;16) AML also decreases p53 acetylation, thus mitigating p53 transcriptional activity, which probably yields the downregulation of several p53-related microRNAs, such as miR-34a.82,83
In CML-Ph1+ blast-phase leukemia wtp53 inactivation is mediated by p53 deacetylation via SIRT1 (HDAC-III) upregulation and by MDM2 upregulation via BCR-ABL–induced La-antigen overexpression.68,84 Another inactivating mechanism is operated by MOZ-TIF2 of inv(8) AML, which causes CBP mislocalization from PML bodies, depletion of its cellular levels, and reduction of CBP-dependent p53 transcriptional activity.85 Variously, AML1-ETO of t(8;21) AML yielded contradicting results as to its effects on wtp53.86,87
AML with deregulated signaling pathways
Aberrant signaling suppresses wtp53 function by various interrelated mechanisms. Aberrantly activated PI3K/AKT and RAS/RAF/MEK/ERK pathways, which are present in most AML cases, yield wtp53 dysfunction mainly via MDM2 activation and by interacting with NF-κB and Wnt/β-catenin pathways.66,88 Importantly, the RAS/RAF/MEK/ERK pathway controls MDM2 expression by transcriptional activation and by promoting MDM2-mRNA nuclear export. This impacts p53 subcellular localization and negatively affects transcription-dependent and independent p53-mediated apoptosis.66,89
Likewise, the constitutively activated NF-κB pathway, which is frequently detected in AML, mediates wtp53 dysfunction by upregulating MDM2-mRNA and protein levels. Notably, wtp53 dysfunction yielded by the above-mentioned deregulated pathways is augmented by AML-related attenuated ARF, which normally counteracts the negative impact of MDM2 and NF-κB on wtp53 levels and function.50,67,90 Likewise, ARF attenuation has a key role in wtp53 functional deficiency, which might accompany the AML-related aberrantly activated Wnt/β-catenin pathway. Normally, beyond its oncogenic effects, hyperactive β-catenin also induces ARF-mediated p53 activation, which is apparently impaired in the face of a mitigated ARF.91,92
Silenced Notch signaling, which is frequently found in AML, is also implicated in wtp53 attenuation. Indeed, Notch agonists were shown to enhance p53 expression along with BCL2 downregulation and growth inhibition of AML cells.93
AML with deregulated miRs
Wtp53 expression might be suppressed by aberrant expression of certain miRs. miR-125b, a p53 negative regulator, is overexpressed in several AML entities, including FLT3-ITD AML.94 By binding to the p53-mRNA 3′ untranslated region, it represses p53 protein levels and function. Additionally, miR-125b represses several targets in p53 network, including apoptosis and cell cycle regulators.95
miR-3151, another p53 negative regulator, is overexpressed in elderly patients with de novo cytogenetically normal AML, serving as an independent poor prognostic factor. By binding to the p53-mRNA 3′ untranslated region, it disrupts p53-mediated apoptosis in AML cells.96 Conversely, miR-29 family operates normally as a p53 positive regulator, reducing PI3K/AKT and MDM2 activity. Downregulation of miR-29b in AML abrogates p53 function, while its restoration revives it.97,98
AML with CRM1 overexpression
Overexpressed CRM1 enhances MDM2-mediated wtp53 nuclear export and cytoplasmic sequestration.99 wtp53 inactivation might be further augmented by CRM1-mediated nuclear export of additional cargo proteins, including NPM1. This molecular setup adds an example for dysfunctional wtp53 associated with adverse cytogenetics and poor prognosis.100
p53-targeted therapy for AML
Despite growth in understanding AML pathobiology, therapeutic progress is still inadequate. The required improvement has yielded development of novel drugs targeting various molecularly defined AML entities, including p53-based therapies.
The encouraging results of clinical trials with MDM2 inhibitors, which address dysfunctional wtp53 protein, and with the hypomethylating decitabine, which decreases mutp53 levels, substantiate the concept that p53-based therapies might improve AML outcome.7,101 Recognizing the extended repertoire of p53 GOF mutants and dysfunctional wtp53 proteins enables identifying p53-based druggable targets and improving patient-oriented therapeutic decisions.12,13,102
Targeting mutp53 in AML
Mutp53 targeted therapy aims to abolish mutp53 cancer cells or to rescue p53 mutational inactivation. The pharmacological strategies are directed toward regaining wtp53-like conformation and tumor-suppressive functions to mutp53 (Table 1, “Reconforming and reactivating drugs”), abrogating distinct mechanisms underlying mutp53 GOF, and promoting mutp53 degradation [Table 1, “Drugs abrogating mutp53 GOF (to be added to RAD)”]. Additionally, several mutp53 RADs modulate the cellular redox state, thus triggering ROS-induced p53-dependent cell-death.12,103
Drugs targeting mutp53 in AML
| . | Drug targets . | Mechanism of action and comments . | AML studies . | Reference . |
|---|---|---|---|---|
| Reconforming and reactivating drugs | ||||
| PRIMA-1MET (APR-246) | Various mutp53 (eg, R273H, R175H) | Converts to MQ, which binds to mutp53 core domain cysteines, thus restoring p53 DNA-binding and apoptosis; induces ROS formation, which augments the cell-death effect | Yes | 12, 104, 105 |
| PK7088/PhiKan083 and other novel Y220C targeting compounds | Y220C (rare in AML, might precede t-AML) | Binds to the mutation-induced surface crevice of mutp53 and stabilizes it; triggers BAX nuclear export to the mitochondria, thus restoring p53 nontranscriptional apoptosis | Preclinical | 115, 20 |
| NSC319726 (ZMC1) | R175H non–zinc binding and other mutp53 with impaired zinc binding | Zinc chelator, providing optimal zinc concentration for mutp53 proper folding; induces ROS formation | Preclinical | 103 |
| Reactivating peptides | Various mutp53 | Bind to the wtp53 conformation of the mutp53 protein, thus shifting the balance toward the wtp53 conformation | Preclinical | 102 |
| Drugsabrogating mutp53 GOF (to be added to RAD) | ||||
| HSP90 inhibitorsTanespimycin (geldanamycin derivative)Ganetespib | mutp53-HSP90 binding | Bind directly with high affinity to the tumor HSP90 ATP-binding pocket; promote MDM2/CHIP E3 ligases-mediated mutp53 degradation; inhibit PI3K-AKT and NF-κB signaling | YesYes | 43, 44, 116www.clinicaltrials.gov #NCT00098423www.clinicaltrials.gov #NCT00858572 |
| Vorinostat HDAC6 inhibitor | Inhibits HSP90 deacetylation and activity | Yes | www.clinicaltrials.gov #NCT00948064 | |
| RETRA | mutp53-p73 binding | Disrupts mutp53-p73 interaction; increases p73 levels; restores p73-mediated expression of p53 target genes (provided p73 is not dysfunctional due to p73 promoter methylation, or aberrant TAp73/ΔNp73 ratio) | Preclinical | 111, 112 |
| MDR modulators: fourth-generation MDR reversal agents and PRIMA-1MET(APR-246) | mutp53-induced MDR1 transactivation | Currently existing MDR1 inhibitors have failed clinically because of systemic toxicity or inefficiency; mutp53 reactivation has an added effect on MDR1 upregulation since wtp53 represses MDRl promoter activity | Preclinical | 47, 113, 117 |
| NF-κB inhibitorsBortezomib proteasome inhibitor | mutp53-mediated NFκB activation (TNFα-induced) and mutp53–NF-κB functional synergism | Inhibit NF-κB nuclear translocation and activity. Selectively target LSCs (in which NF-κB activity is aberrantly increased compared with normal HPSCs). Since NF-κB also enhances MDR1, its inhibition might also address chemoresistance. Beware: full reactivation of mutp53 should precede proteasome-inhibitor treatment | Yes | 50, 51, 118 50 |
| . | Drug targets . | Mechanism of action and comments . | AML studies . | Reference . |
|---|---|---|---|---|
| Reconforming and reactivating drugs | ||||
| PRIMA-1MET (APR-246) | Various mutp53 (eg, R273H, R175H) | Converts to MQ, which binds to mutp53 core domain cysteines, thus restoring p53 DNA-binding and apoptosis; induces ROS formation, which augments the cell-death effect | Yes | 12, 104, 105 |
| PK7088/PhiKan083 and other novel Y220C targeting compounds | Y220C (rare in AML, might precede t-AML) | Binds to the mutation-induced surface crevice of mutp53 and stabilizes it; triggers BAX nuclear export to the mitochondria, thus restoring p53 nontranscriptional apoptosis | Preclinical | 115, 20 |
| NSC319726 (ZMC1) | R175H non–zinc binding and other mutp53 with impaired zinc binding | Zinc chelator, providing optimal zinc concentration for mutp53 proper folding; induces ROS formation | Preclinical | 103 |
| Reactivating peptides | Various mutp53 | Bind to the wtp53 conformation of the mutp53 protein, thus shifting the balance toward the wtp53 conformation | Preclinical | 102 |
| Drugsabrogating mutp53 GOF (to be added to RAD) | ||||
| HSP90 inhibitorsTanespimycin (geldanamycin derivative)Ganetespib | mutp53-HSP90 binding | Bind directly with high affinity to the tumor HSP90 ATP-binding pocket; promote MDM2/CHIP E3 ligases-mediated mutp53 degradation; inhibit PI3K-AKT and NF-κB signaling | YesYes | 43, 44, 116www.clinicaltrials.gov #NCT00098423www.clinicaltrials.gov #NCT00858572 |
| Vorinostat HDAC6 inhibitor | Inhibits HSP90 deacetylation and activity | Yes | www.clinicaltrials.gov #NCT00948064 | |
| RETRA | mutp53-p73 binding | Disrupts mutp53-p73 interaction; increases p73 levels; restores p73-mediated expression of p53 target genes (provided p73 is not dysfunctional due to p73 promoter methylation, or aberrant TAp73/ΔNp73 ratio) | Preclinical | 111, 112 |
| MDR modulators: fourth-generation MDR reversal agents and PRIMA-1MET(APR-246) | mutp53-induced MDR1 transactivation | Currently existing MDR1 inhibitors have failed clinically because of systemic toxicity or inefficiency; mutp53 reactivation has an added effect on MDR1 upregulation since wtp53 represses MDRl promoter activity | Preclinical | 47, 113, 117 |
| NF-κB inhibitorsBortezomib proteasome inhibitor | mutp53-mediated NFκB activation (TNFα-induced) and mutp53–NF-κB functional synergism | Inhibit NF-κB nuclear translocation and activity. Selectively target LSCs (in which NF-κB activity is aberrantly increased compared with normal HPSCs). Since NF-κB also enhances MDR1, its inhibition might also address chemoresistance. Beware: full reactivation of mutp53 should precede proteasome-inhibitor treatment | Yes | 50, 51, 118 50 |
The referred clinical trials evaluated the drugs’ therapeutic efficacy in AML, but not necessarily in view of p53 status.
MQ, methylene quinuclidinone; RAD, reactivating drug; RETRA, reactivation of transcriptional reporter activity; ROS, reactive oxygen species.
Reconforming and RADs targeting mutp53, thereby abrogating its oncogenic GOF properties, are in different stages of evaluation. Currently, the most prominent RAD is APR-246, a methylated version of PRIMA-1 that affects various mutp53 proteins, binds to unfolded wtp53, and has already entered clinical trials, including AML trials.104,105 Its mechanism of action, as well as that of other mutp53-targeting drugs, is itemized in Table 1.
Recently, by adopting phage display technology combined with deep sequencing, we identified mutp53 reactivating peptides (Table 1, “Reconforming and reactivating drugs”) that stabilize mutp53 protein in wtp53 conformation, endowing mutp53 with wtp53-like activities in vitro and in vivo, in several xenograft models.102 Since under physiological conditions there is a constant dynamic equilibrium between wtp53 and mutp53 protein conformations, we assume that the peptides bind to the wtp53 conformation of the mutp53 protein, thereby shifting the balance toward the wtp53 form. These peptides may potentially serve as novel agents for cancer treatment,102 not only in patients with TP53 mutations but also in patients with dysfunctional wtp53 protein. We expect that this initial study, performed on solid tumors, will also be applicable to various hematological malignancies, including AML, which presents a high incidence of dysfunctional wtp53 protein.
Targeting dysfunctional wtp53 in AML
Dysfunctional wtp53 targeted therapy aims to rescue wtp53 inactivation by addressing various AML-related wtp53 inactivating mechanisms (Table 2). One such strategy entails MDM2/4 inhibitors that disrupt wtp53–MDM2/4 interactions.66,106 wtp53’s interaction with MDM2 is mediated by its 15-residue α-helical transactivation domain, which inserts into a hydrophobic binding pocket on the MDM2 surface, primarily through 3 amino acids: Phe19, Trp23, and Leu26.13
Drugs Targeting Dysfunctional wtp53 in AML
| Drugs . | Targeted inactivating mechanisms . | Mechanism of action and comments . | AML studies . | Reference . |
|---|---|---|---|---|
| MDM2 inhibitors | MDM2 upregulation | Inhibit MDM2–p53 interaction, thus promoting p53 stabilization and activation. Efficacy depends on p53 protein/network normalcy. Warning: MDM2 inhibition has the potential to increase the levels of GOF mutp53. | 13 | |
| Nutlins and other sm | The prototype MDM2 antagonists which disrupt wtp53-MDM2 interaction, by binding MDM2 in the p53-binding pocket | No | 13 | |
| Nutlin 3 (sm) (cis-imidazoline) | A research model molecule. Also disrupts the MDM2-p73/E2Fl interaction, thus promoting p53 - independent apoptosis and differentiation. Efficacy in AML is closely related to multiple synergistic interactions with doxorubicin, cytarabine, valproate, AZD6244 (RAF/MEK/ERK inhibitor), PI-103 (PI3/AKT/mTOR inhibitor), sorafenib (FLT3 inhibitor), MK-0457 (pan-Aurora kinase inhibitor), XIAP antisense oligonucleotide (XIAP inhibitor), and ABT-737 (BcL-2 inhibitor). Warning: acquisition of mutp53 in response to Nutlin 3 has been reported in cancer cells. | 13, 107, 111, 119, 120 | ||
| RG7112 (RO5045337) (Nutlin type) | Preliminary clinical trial data (supplemental Table 2) | Yes | 101 | |
| www.clinicaltrials.gov #NCT00623870 | ||||
| NCT01635296 | ||||
| RG7388, RO5503781 Idasanutlin (pyrrolidine class) | Preliminary clinical trial data (supplemental Table 2) | Yes | ||
| www.clinicaltrials.gov #NCT01773408 | ||||
| www.clinicaltrials.gov #NCT02545283 | ||||
| www.clinicaltrials.gov #NCT02670044 | ||||
| RG7775, R06839921 (Pegylated prodrug of Idasanutlin) | Preliminary clinical trial data (unpresented) | Yes | www.clinicaltrials.gov #NCT02098967 | |
| HDM201 (imidazolopyrrolidinone analog) | Preliminary clinical trial data (unpresented) | Yes | www.clinicaltrials.gov #NCT02143635, 121 | |
| AMG 232 (piperidinone) | Preliminary clinical trial data (unpresented) | Yes | www.clinicaltrials.gov #NCT02016729 | |
| www.clinicaltrials.gov #NCT03041688 | ||||
| MK-8242 (sm)* | Preliminary clinical trial data (supplemental Table 2) | Yes | www.clinicaltrials.gov #NCT01451437 | |
| DS-3032b (imidazothiazole) | Preliminary clinical trial data (supplemental Table 2) | Yes | www.clinicaltrials.gov #NCT02319369, 122 | |
| Stapled p53 peptidesALRN-6924 | MDM2/4 upregulation | Dual inhibition of MDM2 and MDM4 and escape resistance to MDM2 inhibition due to upregulated MDM4 | Yes | 13www.clinicaltrials.gov #NCT02909972 |
| RNA Pol I inhibitors | Trigger “nucleolar stress,” yielding p53 activation (which further promotes Pol I repression via inhibition of UBF-SL1 interaction); although wtp53 AML cells are most sensitive to Pol I inhibition, in the face of wtp53 dysfunction, this sensitivity should be reassessed | Yes | 123 | |
| CX-5461 | ACTRN12613001061729† | |||
| HDAC inhibitors | wtp53 de-acetylation (due to overexpressed or aberrantly recruited HDACs) | Acetylation is mutually exclusive with ubiquitylation; reacetylation reactivates wtp53 and sensitizes leukemic cells to MDM2 inhibitors, TKIs, and chemotherapy | Yes | 74, 84 |
| Valproic acid and entinostat (HDAC class l inhibitors) | www.clinicaltrials.gov #NCT00339196 and #NCT01305499 | |||
| Vorinostat and panobinostat (pan-HDAC inhibitors) | Yes | www.clinicaltrials.gov #NCT00948064 and #NCT01613976 | ||
| Tenovin 6 SIRT1/2 (HDAC class III) inhibitor | Research molecule | No | 124 | |
| SIRT1/2 inhibitors | Under development; although FLT3-ITD AML cells are sensitive to SIRT1 inhibitors, their efficacy might be limited as FLT3-ITD is considered a cooperating event in AML | No | 124 | |
| SINEs | CRM1 overexpression | Inhibit the CRM1 exporter; redirect wtp53 to the nucleus, thus promoting its transcriptional activities | 100 | |
| KPT-185 | KPT-185 also affects p53 by synergizing with Nutlin 3 in p53 nuclear retention, promoting NPMlc+ nucleolar relocalization and downregulating FLT3 expression | No | ||
| Selinexor (KPT-330) | Yes | www.clinicaltrials.gov #NCT02403310, #NCT02093403, #NCT02088541, #NCT02299518, #NCT02530476, #NCT02485535, and #NCT02573363 |
| Drugs . | Targeted inactivating mechanisms . | Mechanism of action and comments . | AML studies . | Reference . |
|---|---|---|---|---|
| MDM2 inhibitors | MDM2 upregulation | Inhibit MDM2–p53 interaction, thus promoting p53 stabilization and activation. Efficacy depends on p53 protein/network normalcy. Warning: MDM2 inhibition has the potential to increase the levels of GOF mutp53. | 13 | |
| Nutlins and other sm | The prototype MDM2 antagonists which disrupt wtp53-MDM2 interaction, by binding MDM2 in the p53-binding pocket | No | 13 | |
| Nutlin 3 (sm) (cis-imidazoline) | A research model molecule. Also disrupts the MDM2-p73/E2Fl interaction, thus promoting p53 - independent apoptosis and differentiation. Efficacy in AML is closely related to multiple synergistic interactions with doxorubicin, cytarabine, valproate, AZD6244 (RAF/MEK/ERK inhibitor), PI-103 (PI3/AKT/mTOR inhibitor), sorafenib (FLT3 inhibitor), MK-0457 (pan-Aurora kinase inhibitor), XIAP antisense oligonucleotide (XIAP inhibitor), and ABT-737 (BcL-2 inhibitor). Warning: acquisition of mutp53 in response to Nutlin 3 has been reported in cancer cells. | 13, 107, 111, 119, 120 | ||
| RG7112 (RO5045337) (Nutlin type) | Preliminary clinical trial data (supplemental Table 2) | Yes | 101 | |
| www.clinicaltrials.gov #NCT00623870 | ||||
| NCT01635296 | ||||
| RG7388, RO5503781 Idasanutlin (pyrrolidine class) | Preliminary clinical trial data (supplemental Table 2) | Yes | ||
| www.clinicaltrials.gov #NCT01773408 | ||||
| www.clinicaltrials.gov #NCT02545283 | ||||
| www.clinicaltrials.gov #NCT02670044 | ||||
| RG7775, R06839921 (Pegylated prodrug of Idasanutlin) | Preliminary clinical trial data (unpresented) | Yes | www.clinicaltrials.gov #NCT02098967 | |
| HDM201 (imidazolopyrrolidinone analog) | Preliminary clinical trial data (unpresented) | Yes | www.clinicaltrials.gov #NCT02143635, 121 | |
| AMG 232 (piperidinone) | Preliminary clinical trial data (unpresented) | Yes | www.clinicaltrials.gov #NCT02016729 | |
| www.clinicaltrials.gov #NCT03041688 | ||||
| MK-8242 (sm)* | Preliminary clinical trial data (supplemental Table 2) | Yes | www.clinicaltrials.gov #NCT01451437 | |
| DS-3032b (imidazothiazole) | Preliminary clinical trial data (supplemental Table 2) | Yes | www.clinicaltrials.gov #NCT02319369, 122 | |
| Stapled p53 peptidesALRN-6924 | MDM2/4 upregulation | Dual inhibition of MDM2 and MDM4 and escape resistance to MDM2 inhibition due to upregulated MDM4 | Yes | 13www.clinicaltrials.gov #NCT02909972 |
| RNA Pol I inhibitors | Trigger “nucleolar stress,” yielding p53 activation (which further promotes Pol I repression via inhibition of UBF-SL1 interaction); although wtp53 AML cells are most sensitive to Pol I inhibition, in the face of wtp53 dysfunction, this sensitivity should be reassessed | Yes | 123 | |
| CX-5461 | ACTRN12613001061729† | |||
| HDAC inhibitors | wtp53 de-acetylation (due to overexpressed or aberrantly recruited HDACs) | Acetylation is mutually exclusive with ubiquitylation; reacetylation reactivates wtp53 and sensitizes leukemic cells to MDM2 inhibitors, TKIs, and chemotherapy | Yes | 74, 84 |
| Valproic acid and entinostat (HDAC class l inhibitors) | www.clinicaltrials.gov #NCT00339196 and #NCT01305499 | |||
| Vorinostat and panobinostat (pan-HDAC inhibitors) | Yes | www.clinicaltrials.gov #NCT00948064 and #NCT01613976 | ||
| Tenovin 6 SIRT1/2 (HDAC class III) inhibitor | Research molecule | No | 124 | |
| SIRT1/2 inhibitors | Under development; although FLT3-ITD AML cells are sensitive to SIRT1 inhibitors, their efficacy might be limited as FLT3-ITD is considered a cooperating event in AML | No | 124 | |
| SINEs | CRM1 overexpression | Inhibit the CRM1 exporter; redirect wtp53 to the nucleus, thus promoting its transcriptional activities | 100 | |
| KPT-185 | KPT-185 also affects p53 by synergizing with Nutlin 3 in p53 nuclear retention, promoting NPMlc+ nucleolar relocalization and downregulating FLT3 expression | No | ||
| Selinexor (KPT-330) | Yes | www.clinicaltrials.gov #NCT02403310, #NCT02093403, #NCT02088541, #NCT02299518, #NCT02530476, #NCT02485535, and #NCT02573363 |
The referred clinical trials were evaluated as to the treatment effects on AML, but not necessarily with respect to p53 status.
sm, small molecules; SINEs, selective inhibitors of nuclear export; TKIs, tyrosine kinases inhibitors; UBF-SL1, upstream binding factor-selectivity factor 1
Chemical structure has not been disclosed regarding MK-8242.
Australian New Zealand Clinical Trials Registry trial ID.
Several classes of small molecules that selectively bind to MDM2 by mimicking these amino acids and activate wtp53 in a nongenotoxic manner have been advanced into AML clinical studies.106
They differ in their chemical structures, pharmacokinetics, MDM2-binding affinities, and therapeutic index (Table 2, “MDM2 inhibitors,” and supplemental Table 2). Several MDM2i’s capture additional interactions, thus achieving MDM2-binding affinities that are >1000 times higher than those of wtp53 peptides.106
Currently, MDM2i are evaluated as monotherapy and in combination with Cytosar and molecularly targeted agents (venetoclax, trametinib, and decitabine). As monotherapy, MDM2i shows preliminary modest activity.
A major concern of MDM2i therapy is its on-target toxicity to normal cells due to MDM2i-induced p53 activation, to which the hematopoietic and gastrointestinal systems are particularly sensitive.
Another concern refers to the acquisition or selection of TP53 mutations during MDM2i therapy.21,107 Such an appearance, whose significance is to be elucidated, might be a potential mechanism of MDM2i resistance. Additional resistance mechanisms include high MDM4 levels and possible point mutations in the p53-binding pocket of MDM213,108 (Table 2, “MDM2 inhibitors,” and supplemental Table 2). The long-term and off-target toxicities of MDM2i still need to be evaluated.
Though the principle features of wtp53 binding to MDM2 are preserved in its interaction with MDM4, the structural differences in the MDM4 N-terminal p53-binding pocket yield a low-affinity binding to MDM2 inhibitors.66 Since enhanced functionality of wtp53 necessitates concomitant inhibition of MDM2 and MDM4, targeted MDM4 inhibition is required. Indeed, several stabilized stapled α-helical peptides, which compete for the α-helical–mediated wtp53 binding to MDM2 and to MDM4 and escape MDM4-mediated resistance to MDM2 inhibition, are currently under evaluation. In preclinical studies, they displayed increased helicity, protease resistance, and enhanced biological potency.13,109 The newest stapled peptide, ALRN-6924, has already entered a phase 1 AML clinical trial (Table 2, “MDM2 inhibitors”).
Additional potential strategies for rebooting dysfunctional wtp53 address several other p53 modulators, namely RNA polymerase I, HDACs I/III, and nuclear export proteins (Table 2, “RNA Pol-I inhibitors,” “HDAC inhibitors,” and “SINEs”). Some of these compounds have already been evaluated in AML clinical trials, though not in view of p53 status.
p53-based diagnostic workup in AML: a promising future approach
The proposed analysis of p53 status and its potential future therapeutic implications are presented in Figure 4. The noted genotyping methods have already become routine practice, while the phenotyping methods are currently used only for research and clinical trials purposes.110 However, recognizing the significant involvement of dysfunctional p53 in AML will eventually prompt future implementation of p53 functional diagnostics into clinical workup as part of a comprehensive diagnostic approach aiming to improve personalized therapeutic decisions. Accordingly, the following suggested therapies merely represent a conceptual model for future potential therapeutic strategies.
Patient-specific evaluation of p53 status: projection on future possible targeted therapy. Cytogenetics and fluorescence in situ hybridization (FISH) detect 17p− (associated with mutp53 and LOH). SNP-As (single-nucleotide polymorphism arrays), identify CNAs and LOH at a deeper level of detection than conventional cytogenetics and are also used to detect common polymorphisms, including those of TP53. The Sanger sequencing method continues to be a mainstay technology for rapid analysis and sequence determination of relatively small fragments of human DNA, embracing single or few genes at a time. Despite the relatively low of sensitivity of the method (VAF detection lower than ∼20%), it is still considered the gold standard for mutation analysis and is commonly used to confirm NGS results. NGS technology is the current gold standard for comprehensive DNA variant analysis. It enables rapid sequencing of large segments of an individual’s DNA and facilitates precise detection of genetic variations ranging from single-nucleotide substitution to large structural rearrangement, with a VAF detection sensitivity of 2% to 5%. It is based upon preparation of DNA fragment libraries, which are subsequently clonally amplified and sequenced by synthesis in multiple parallel reaction. The generated sequences are aligned and assembled on a human reference genome and computationally analyzed. The most commonly used target enrichment techniques include hybrid capture of target genes or multiplex-based PCR.17,125 NGS assays (selected either for whole-genome/exome/target sequencing based upon the AML gene panel) identify TP53 mutations and related genomic variations, including LOH and CNAs, and provide quantitative measurement of TP53 mutation abundance. Calculating TP53 mutation variant allele frequency allows one to distinguish an initiator mutation from a cooperating one. Additionally, appropriate bioinformatic methods allow SNP calling from NGS data, enabling distinction between a consequential variant and an inconsequential SNP.126 Distinction between somatic and germ line mutations necessitates availability of a matched control tissues sample (eg, patient’s skin cells), which is presumed to retain a germ line configuration. Tag-based methods detect ultrarare TP53 mutations.127 p53 protein expression and activity should be evaluated independently of p53 mutational status. p53 protein levels, assessed by western blotting and immunohistochemistry, should be interpreted cautiously, since p53 overexpression is not a surrogate for TP53 mutations (might be caused by numerous nonmutational stimuli), while lack of expression does not rule out their presence (eg, in cases of TP53 nonsense mutations/deletions).6,128 p53 activity can be evaluated by in vitro (AML blasts) assessment of p53 and its target genes before and after exposure to genotoxic stress induced by Ara-C or anthracyclines. The preferable target genes are p21 and PUMA, whose activation results normally in cell cycle arrest/senescence and apoptosis. These effects are usually assessed by measuring protein levels and by cell cycle/apoptosis and cell viability assays. The HL-60 cell line, characterized by major TP53 deletions, serves as the negative control for p53 functional evaluation.9 A discrepancy between wtp53 expression and an attenuated genotoxic stress response can be attributed to wtp53 inactivation. In case of mutp53, no stress response can be attributed to mutp53 LOF or the presence of mutant p53 GOF, which requires further assessment. The suggested therapies represent merely a conceptual model for future potential therapeutic strategies. In Red: current practice and agents that are already in AML clinical trials with reference to p53 status. *With the exception of RETRA and MDR reversal agents (italicized), all the agents mentioned are already in AML clinical trials, though not with reference to p53 status. Pretreatment ex vivo drug sensitivity testing of patient-derived AML cells may predict efficacy of the suggested therapeutic regimen. Pol I inhibitors, which augment p53 activity, are not listed herein, since their efficacy in AML with mutp53 or inactivated wtp53 is currently unknown.
Patient-specific evaluation of p53 status: projection on future possible targeted therapy. Cytogenetics and fluorescence in situ hybridization (FISH) detect 17p− (associated with mutp53 and LOH). SNP-As (single-nucleotide polymorphism arrays), identify CNAs and LOH at a deeper level of detection than conventional cytogenetics and are also used to detect common polymorphisms, including those of TP53. The Sanger sequencing method continues to be a mainstay technology for rapid analysis and sequence determination of relatively small fragments of human DNA, embracing single or few genes at a time. Despite the relatively low of sensitivity of the method (VAF detection lower than ∼20%), it is still considered the gold standard for mutation analysis and is commonly used to confirm NGS results. NGS technology is the current gold standard for comprehensive DNA variant analysis. It enables rapid sequencing of large segments of an individual’s DNA and facilitates precise detection of genetic variations ranging from single-nucleotide substitution to large structural rearrangement, with a VAF detection sensitivity of 2% to 5%. It is based upon preparation of DNA fragment libraries, which are subsequently clonally amplified and sequenced by synthesis in multiple parallel reaction. The generated sequences are aligned and assembled on a human reference genome and computationally analyzed. The most commonly used target enrichment techniques include hybrid capture of target genes or multiplex-based PCR.17,125 NGS assays (selected either for whole-genome/exome/target sequencing based upon the AML gene panel) identify TP53 mutations and related genomic variations, including LOH and CNAs, and provide quantitative measurement of TP53 mutation abundance. Calculating TP53 mutation variant allele frequency allows one to distinguish an initiator mutation from a cooperating one. Additionally, appropriate bioinformatic methods allow SNP calling from NGS data, enabling distinction between a consequential variant and an inconsequential SNP.126 Distinction between somatic and germ line mutations necessitates availability of a matched control tissues sample (eg, patient’s skin cells), which is presumed to retain a germ line configuration. Tag-based methods detect ultrarare TP53 mutations.127 p53 protein expression and activity should be evaluated independently of p53 mutational status. p53 protein levels, assessed by western blotting and immunohistochemistry, should be interpreted cautiously, since p53 overexpression is not a surrogate for TP53 mutations (might be caused by numerous nonmutational stimuli), while lack of expression does not rule out their presence (eg, in cases of TP53 nonsense mutations/deletions).6,128 p53 activity can be evaluated by in vitro (AML blasts) assessment of p53 and its target genes before and after exposure to genotoxic stress induced by Ara-C or anthracyclines. The preferable target genes are p21 and PUMA, whose activation results normally in cell cycle arrest/senescence and apoptosis. These effects are usually assessed by measuring protein levels and by cell cycle/apoptosis and cell viability assays. The HL-60 cell line, characterized by major TP53 deletions, serves as the negative control for p53 functional evaluation.9 A discrepancy between wtp53 expression and an attenuated genotoxic stress response can be attributed to wtp53 inactivation. In case of mutp53, no stress response can be attributed to mutp53 LOF or the presence of mutant p53 GOF, which requires further assessment. The suggested therapies represent merely a conceptual model for future potential therapeutic strategies. In Red: current practice and agents that are already in AML clinical trials with reference to p53 status. *With the exception of RETRA and MDR reversal agents (italicized), all the agents mentioned are already in AML clinical trials, though not with reference to p53 status. Pretreatment ex vivo drug sensitivity testing of patient-derived AML cells may predict efficacy of the suggested therapeutic regimen. Pol I inhibitors, which augment p53 activity, are not listed herein, since their efficacy in AML with mutp53 or inactivated wtp53 is currently unknown.
Detection of mechanisms underlying mutp53 GOF is promoted in mutp53-AML with chromosomal and amplification instability and aims to identify those with therapeutic relevance (Table 1). mutp53-p73 binding can be assessed by analyzing p21 and PUMA expression following induction by anthracyclines and Nutlin-3 (which does not effect mutp53). RETRA-enforced induction indicates disruption of mutp53-p73 binding and predicts potential efficacy of RETRA treatment.111,112 mutp53-HSP90 binding can be appraised by evaluating mutp53 protein levels and ubiquitination status and activity, before and after exposure to HSP90 inhibitors, whose potential therapeutic benefit might thus be predicted.43,44 Mutp53-induced MDR1 upregulation increases P-gp expression, which can be evaluated on the myeloblast surface. Although fourth-generation MDR reversal agents are currently not available, detection of MDR1 upregulation is still warranted, as it fosters RAD treatment, which downregulates MDR1 by restoring wtp53-like activity to mutp53.113 Mutp53-mediated NF-κB activation is established by chromatin immunoprecipitation, demonstrating concomitant recruitment of mutp53 and NF-κB on target promoters.49
For such AML cases, RADs and NF-κB inhibitors might be beneficial, provided that full reactivation of mutp53 precedes proteasome-inhibitor (NF-κBi) treatment.
Detection of mechanisms underlying wtp53 dysfunction is also of potential therapeutic value (Table 2). MDM2 overexpression can be detected by measuring MDM2 protein levels, and assessing Nutlin 3–induced p53 activation. In such a case, MDM2 inhibitors will probably be beneficent. MDM2 overexpression due to activated signaling pathways might be addressed by adding targeted agents (eg, inhibitors of MEK, mutated FLT3 receptor, and NF-κB), which have already been tested in clinical trials. Being a major negative p53 regulator, MDM2 inhibitors might be effective in AML with dysfunctional wtp53, regardless of the specific inactivating mechanism. Deacetylated p53 and SIRT1 overexpression can be detected by immunoblotting.74,84 In cases of non–SIRT1-mediated deacetylation, HDAC and MDM2 inhibitors might be beneficial. wtp53 cytoplasmic sequestration, confirmed by immunocytochemical staining,114 can be reversed by CRM1 inhibition.
The apoptotic effect might be further augmented by MDM2 inhibitor, which promotes p53 nuclear localization and activity.100
Low ARF expression secondary to methylation can be addressed by demethylating agents, while low expression caused by NPMc+ might be targeted by CRM1 inhibitor, which restores NPMc+ and ARF nucleolar localization and function. MDM2 inhibitors might be of added therapeutic value as ARF attenuation enhances MDM2 activity.
Conclusions
p53 has a major impact on AML evolution, therapy response, and prognosis. Additionally, it centers a complex web of biological interactions, which are abrogated in AML, leaving distinctive fingerprints in the various AML entities.
Currently, in addition to the rather low TP53 mutation rate in AML, the frequent occurrence of dysfunctional wtp53 is being unveiled.
The impact of the latter on AML biology and prognosis might be of greater magnitude and significance than is currently appreciated.
In the era of personal diagnostics and treatments, disclosing the diversity of the various p53 mutants and dysfunctional wtp53 proteins in the individual AML patient operates in line with the goal of precision medicine. Integrating p53 genotyping and functional phenotyping to the routine diagnostic AML workup might yield valuable information for AML subcategorization, prognostication, and selection of the most suitable individualized p53-based treatment.
The online version of this article contains a data supplement.
Acknowledgments
The authors acknowledge the support by the Center of Excellence of the Flight-Attendant Medical Research Institute (FAMRI), the Israel Science Foundation ISF-MOKED Center, the Israeli Academy of Science, and Israel Cancer Research Funds. V.R. is the incumbent of the Norman and Helen Asher Professorial Chair for Cancer Research at the Weizmann Institute.
Authorship
Contribution: V.R. and M.P. conceived the idea for the review and the proposed concept and contributed equally to the literature search, figures, writing, and critical review of the manuscript; and A.M. contributed to the preparation and editing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Varda Rotter, Department of Molecular Cell Biology, The Weizmann Institute of Science, Rehovot 76100, Israel; e-mail: varda.rotter@weizmann.ac.il.