Key Points
Platelet MPs infiltrate solid tumors and transfer platelet-derived miRNAs to tumor cells within solid tumors in vivo.
Transfer of platelet miRNAs to tumor cells results in downregulation of tumor cell genes and inhibition of solid tumor growth.
Abstract
Platelet-derived microparticles (PMPs) are associated with enhancement of metastasis and poor cancer outcomes. Circulating PMPs transfer platelet microRNAs (miRNAs) to vascular cells. Solid tumor vasculature is highly permeable, allowing the possibility of PMP–tumor cell interaction. Here, we show that PMPs infiltrate solid tumors in humans and mice and transfer platelet-derived RNA, including miRNAs, to tumor cells in vivo and in vitro, resulting in tumor cell apoptosis. MiR-24 was a major species in this transfer. PMP transfusion inhibited growth of both lung and colon carcinoma ectopic tumors, whereas blockade of miR-24 in tumor cells accelerated tumor growth in vivo, and prevented tumor growth inhibition by PMPs. Conversely, Par4-deleted mice, which had reduced circulating microparticles (MPs), supported accelerated tumor growth which was halted by PMP transfusion. PMP targeting was associated with tumor cell apoptosis in vivo. We identified direct RNA targets of platelet-derived miR-24 in tumor cells, which included mitochondrial mt-Nd2, and Snora75, a noncoding small nucleolar RNA. These RNAs were suppressed in PMP-treated tumor cells, resulting in mitochondrial dysfunction and growth inhibition, in an miR-24–dependent manner. Thus, platelet-derived miRNAs transfer in vivo to tumor cells in solid tumors via infiltrating MPs, regulate tumor cell gene expression, and modulate tumor progression. These findings provide novel insight into mechanisms of horizontal RNA transfer and add multiple layers to the regulatory roles of miRNAs and PMPs in tumor progression. Plasma MP-mediated transfer of regulatory RNAs and modulation of gene expression may be a common feature with important outcomes in contexts of enhanced vascular permeability.
Introduction
Platelets have been associated with tumor progression and metastatic dissemination through platelet–tumor cell (TC) interactions.1-7 Platelet-TC interactions may contribute to tumor progression in several ways, including enhancing cancer-related coagulation and providing a TC “shroud” to shield them from the immune system.8 The presence of cancer increases platelet production, which has been associated with poorer outcomes in multiple cancers.9,10 Platelets can stimulate proliferation of human and murine cancer cells in a manner that does not require platelet-tumor contact.11 However, the platelet-cancer axis still remains unsolved and is an area of active investigation.
Platelets and other cells release microparticles (MPs) into the plasma in response to receptor agonists and shear stress.12 At least 45% of plasma-borne MPs are platelet-derived MPs (PMPs).13,14 PMP release increases in individuals bearing solid tumors, but roles of PMPs in cancer progression are incompletely understood.15,16 PMPs are enriched in platelet microRNAs (miRNAs), a small cohort of which are present at high copy number, accounting for the bulk of plasma miRNAs.17-21 Purified PMPs are able to transfer at least some miRNA content to cells following coincubation in vitro, and regulate gene expression.22-24 Several miRNAs enriched in PMPs, including miR-27a, miR-24, miR-155, miR-195, let-7a/b, and miR-223, target both tumor suppressor genes and oncogenes (in multiple cancer types), have been identified as diagnostic and prognostic markers of malignancy, and have been implicated in therapy resistance.25-46 Tumor neovasculature is highly permeable, which we predicted might allow circulating MPs direct access to tumor cells. In this study, we investigated PMP infiltration in solid tumors, transfer of platelet-derived miRNAs to tumor cells, and cellular and physiological effects.
Methods
Tumor allografts and immune-induced thrombocytopenia
A suspension of 1 × 106 cells/200 μL of Hanks balanced salt solution was injected subcutaneously into the shaved flank of 8-week-old mice. Thrombocytopenia was induced by intraperitoneal (i.p.) injection of 50 mg/kg rat-anti-mouse CD41 antibodies. Platelet count was assessed by Hemavet analysis, and was <20% of starting counts after 24 hours. In some cases, mice were injected i.p. with 100 mg/kg 4-thiouracil (4TU) in 20% dimethyl sulfoxide/80% corn oil, or vehicle. Tumor volume was calculated as described.47 Tumors were resected from euthanized mice, and cleaned of fat, skin, and connective tissue for further processing.
miRNA:messenger RNA target adduct formation and screening
Cells suspended in 500 μL of fractionation buffer (20 mM Tris-Cl pH 7.4,100 mM NaCl, 2 mM MgOAc, 5 mM KCl, protease inhibitor cocktail [Roche, Indianapolis, IN]) were dounce-homogenized followed by sonication. Lysates were treated with 1 U/μL RNase T1 (Fisher, Pittsburgh, PA) for 15 minutes at 22°C, followed by RNase inhibition with 10 mM MnCl2. T4 polynucleotide kinase (PNK) minus 1 U/μL plus 100 mM adenosine triphosphate (ATP), 5 mM dithiothreitol was added (40 minutes at 10°C). RNA ligation was carried out with 0.2 U/μL T4 RNA ligase (16 hours at 4°C). RNA was extracted in TRIzol and resuspended in diethyl pyrocarbonate–treated H2O, followed by 1 μL of PNK and no ATP for 40 minutes at 10°C, and poly(dA) tailing and first-strand complementary DNA (cDNA) synthesis with the NCode kit (see supplemental Methods, available on the Blood Web site).48 cDNA libraries were subjected to Taq polymerase chain reaction (PCR) using miR-specific 5′ oligonucleotides and universal poly(dT) 3′ primers. Products were subcloned by direct ligation of the reaction mixture into the pCR2.1 TA vector (Invitrogen), and ligation reactions were transformed into DH5α Escherichia coli for ampicillin selection, colony propagation, plasmid DNA minipreps (Qiagen, Valencia, CA), and sequencing.
Results
Platelet MPs infiltrate solid tumors
Circulating PMPs harbor miRNAs and can transfer platelet-derived miRNAs to endothelium and leukocytes.22,23,49 Because tumor blood vessels are highly permeable due to endothelial dysfunction and poor pericyte coverage,50 and PMP release correlates with solid tumor growth and metastasis,4,16,51 we considered whether TCs in solid tumors are targets of PMPs. We observed PMP infiltration, indicated by antibodies to αIIb integrin (CD41), a platelet/megakaryocyte-specific receptor and a PMP marker,52 in the extravascular tumor environment as indicated by von Willebrand factor (VWF) staining for blood vessels, in grade II/III solid tumors derived from human patients, but not in adjacent normal tissue, in multiple cancer types (Figure 1A). The puncta ranged in diameter from ∼100 to 1000 nm, the diameter range of PMPs,53,54 and were Annexin V+ (Figure 1B), indicating phosphatidylserine exposure on the outer leaflet, a characteristic of MPs and apoptotic cells. Most, but not all, Annexin V+ puncta in the tumor sections also contained αIIb integrin, consistent with PMPs being the major MP fraction in the infiltrates (Figure 1B). Examination of tissue sections spiked with freshly isolated platelets and stained with αIIb integrin antibodies confirmed that the platelet-derived intratumoral material consisted of platelet fragments smaller than intact platelets (Figure 1C). PMP tumor infiltration was observed across tumor grades in lung and colon cancer subtypes, but extravascular PMPs were not observed in paired, uninvolved normal tissues except for a few cases (Figure 1D-G; Table 1). In these latter cases, PMP infiltration was only evident in normal tissue adjacent to the tumor, suggesting that infiltration reflected a specific effect of proximity to the tumor microenvironment (Figure 1F).
PMP infiltration in solid tumors in human patients. (A) Tissue microarray slides containing 5-μm sections from the indicated human tumors and uninvolved adjacent tissue (“Normal”) were stained with the indicated antibodies and 4′,6-diamidino-2-phenylindole (DAPI). Colon, grade I-II colon carcinoma; lung, grade II lung squamous cell carcinoma; prostate, grade II prostate adenocarcinoma; liver, grade II-III hepatocellular carcinoma; breast, grade II-III invasive ductal carcinoma. αIIb integrin, green; VWF, red; DAPI, blue. Bottom row, center area insets, original magnification ×3. Bars, 50 μm (n = 4). (B) Representative images from panel A, showing counterstain with fluorescein isothiocyanate (FITC)-Annexin V (AXV; shown as red). αIIb integrin, green; DAPI, blue. Merged images with DAPI shown to the right; αIIb integrin/Annexin V overlap appears as yellow. VWF staining was omitted from the merged images for clarity. (C) A section of human lung adenocarcinoma, grade II was incubated with 103 freshly isolated murine platelets for 15 minutes before being fixed and stained as indicated. Yellow arrowheads indicate ectopic intact platelets. (D) Representative images from human lung cancer array with paired uninvolved tissue, stained as in panel A. (E) Representative images from human colon cancer array with paired uninvolved tissue. Note that some αIIb integrin-positive platelets can be seen within VWF-labeled blood vessels. (F) Representative image of colon adenocarcinoma, grade III, including adjacent normal tissue, showing PMP infiltration in the uninvolved tissue adjacent to the tumor border (indicated with a dotted line). Bars (B-F), 25 μm. (G) Percentage of PMP+ tissues from total assayed tissues for colon adenocarcinomas and lung cancers, and adjacent uninvolved tissue, shown ± standard error of the mean (SEM) (n = 3). Colon, P < .01; lung, P < .004. AC, adenocarcinoma; BAC, bronchioalveolar carcinoma; PC, papillary carcinoma; SCC, squamous cell carcinoma; SCLC, small cell lung cancer.
PMP infiltration in solid tumors in human patients. (A) Tissue microarray slides containing 5-μm sections from the indicated human tumors and uninvolved adjacent tissue (“Normal”) were stained with the indicated antibodies and 4′,6-diamidino-2-phenylindole (DAPI). Colon, grade I-II colon carcinoma; lung, grade II lung squamous cell carcinoma; prostate, grade II prostate adenocarcinoma; liver, grade II-III hepatocellular carcinoma; breast, grade II-III invasive ductal carcinoma. αIIb integrin, green; VWF, red; DAPI, blue. Bottom row, center area insets, original magnification ×3. Bars, 50 μm (n = 4). (B) Representative images from panel A, showing counterstain with fluorescein isothiocyanate (FITC)-Annexin V (AXV; shown as red). αIIb integrin, green; DAPI, blue. Merged images with DAPI shown to the right; αIIb integrin/Annexin V overlap appears as yellow. VWF staining was omitted from the merged images for clarity. (C) A section of human lung adenocarcinoma, grade II was incubated with 103 freshly isolated murine platelets for 15 minutes before being fixed and stained as indicated. Yellow arrowheads indicate ectopic intact platelets. (D) Representative images from human lung cancer array with paired uninvolved tissue, stained as in panel A. (E) Representative images from human colon cancer array with paired uninvolved tissue. Note that some αIIb integrin-positive platelets can be seen within VWF-labeled blood vessels. (F) Representative image of colon adenocarcinoma, grade III, including adjacent normal tissue, showing PMP infiltration in the uninvolved tissue adjacent to the tumor border (indicated with a dotted line). Bars (B-F), 25 μm. (G) Percentage of PMP+ tissues from total assayed tissues for colon adenocarcinomas and lung cancers, and adjacent uninvolved tissue, shown ± standard error of the mean (SEM) (n = 3). Colon, P < .01; lung, P < .004. AC, adenocarcinoma; BAC, bronchioalveolar carcinoma; PC, papillary carcinoma; SCC, squamous cell carcinoma; SCLC, small cell lung cancer.
Presence of extravascular PMPs scored in graded lung carcinoma and colon adenocarcinoma, and adjacent uninvolved tissues
| Type and grade . | Lung carcinomas . | Colon adenocarcinomas . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC I . | AC II . | AC III . | BAC . | PC . | SCC I . | SCC II . | SCC III . | SCLC . | AC I-II . | AC II . | AC II-III . | AC III . | AC III-IV . | |
| Tumor | 2/2 | 2/2 | 6/6 | 2/2 | 4/4 | 2/2 | 1/2 | 7/8 | 3/4 | 5/6 | 5/5 | 3/4 | 10/10 | 3/6 |
| Uninvolved paired normal tissue | 0/2 | 1/2 | 0/6 | 0/2 | 1/4 | 0/2 | 1/2 | 0/8 | 0/4 | 1/6 | 1/6 | 0/6 | 0/6 | 1/6 |
| Type and grade . | Lung carcinomas . | Colon adenocarcinomas . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC I . | AC II . | AC III . | BAC . | PC . | SCC I . | SCC II . | SCC III . | SCLC . | AC I-II . | AC II . | AC II-III . | AC III . | AC III-IV . | |
| Tumor | 2/2 | 2/2 | 6/6 | 2/2 | 4/4 | 2/2 | 1/2 | 7/8 | 3/4 | 5/6 | 5/5 | 3/4 | 10/10 | 3/6 |
| Uninvolved paired normal tissue | 0/2 | 1/2 | 0/6 | 0/2 | 1/4 | 0/2 | 1/2 | 0/8 | 0/4 | 1/6 | 1/6 | 0/6 | 0/6 | 1/6 |
Stained slides from Figure 1C-D were scored for intratumoral PMPs by observers blinded to the sample type, and shown as a ratio of PMP+ to total. Samples were counted as negative if no PMPs were observed in 5 separate fields of 500 μm2.
To gain mechanistic insight, we used an ectopic solid tumor allograft model in mice using Lewis lung carcinoma (LLC) cells injected as a bolus subcutaneously into mouse flanks.55 PMPs were observed infiltrating LLC-allografted tumors in mice (Figure 2A), similar to human tumors (Figure 1). CD63, a marker for both PMPs and exosomes,13 overlapped with αIIb integrin in PMP-like structures (CD63-only exosomes were also evident), confirming that the αIIb integrin-positive structures represent PMPs and not αIIb integrin expression in TCs, which has been observed in metastatic melanoma and some cancer cell lines (Figure 2B).56-59 Many TCs isolated from resected tumors at 21 days were decorated with PMPs (62.8% ± 3.2% of TCs; n = 5; >250 cells per experiment), indicating association of infiltrating PMPs with the TCs (Figure 2C). PMPs were also associated with green fluorescent protein–positive (GFP+) TCs isolated from resected tumors from NIH3T3 cells transformed with GFP-HRAS(G12V)47,60 (supplemental Figure 1A). Fewer PMPs appeared associated with TCs derived from 7-day tumors (supplemental Figure 1B), suggesting that robust neovascularization precludes PMP extravasation in solid tumors. TCs extracted at day 21, 24 hours after immune-induced thrombocytopenia, showed substantially reduced PMPs (3.1% ± 2.2% of TCs), indicating that the MPs were derived from blood platelets (supplemental Figure 1C). Many of the PMPs were attached to TC surfaces, and αIIb integrin could also be seen internalized in HRAS-containing recycling endosomes, suggesting PMP cargo internalization (supplemental Figure 1D).61,62 In some cells, αIIb integrin was broadly distributed at the plasma membrane, indicating redistribution of PMP-derived proteins in the TCs, either as a result of plasma membrane fusion, transport of internalized protein to the target cell membrane, or expression of platelet-derived messenger RNA (mRNA) (supplemental Figure 1E). Thus, platelet MPs can undergo extravasation beyond the blood circulation in solid tumors and associate with tumor cells.
PMP infiltration in solid tumor allografts and RNA transfer in mice. (A) Immunohistochemistry (IHC) in a 5-μm section from LLC tumor allograft, 21 days. αIIb integrin, red. (B) IHC in a 5-μm section from LLC tumor allograft, 21 days. αIIb integrin, red (top); CD63, green (middle); DAPI, blue (shown in merged image, bottom). αIIb integrin/CD63 overlap appears in some areas in the merged image as yellow. (C) Tumor cells from a resected LLC tumor at 21 days, isolated as described in supplemental Methods. αIIb integrin, red; DAPI, blue. (D-E) Tumor cells from resected LLC allograft 24 hours after transfusion of AO-labeled platelets. (D) αIIb integrin, red; (E) AO, green. Bars (A-E), 10 μm. *PMP− cells. (F) Percentage of cells with cytosolic AO staining, shown ± SEM (n = 3, >100 cells each). (G) Human platelets were transfected with unlabeled siRNA and transfused into mice bearing 20-day LLC tumors. After 24 hours, tumors were resected, digested, cleared of vascular cells with α-CD31 beads, and tumor cells were captured on fibronectin-coated coverslips. (H) Tumor cells ex vivo as in panel G, from mice transfused with human platelets transfected with FAM-siRNA. (I) LLC cells, treated in vitro with PMPs derived from human platelets 48 hours after platelet transfection with unlabeled siRNA. (J) Cells treated as in panel I with PMPs derived from human platelets transfected with FAM-siRNA. (H-J) Human αIIb integrin, red; FAM, green; DAPI, blue. Dashed white lines indicate cell borders as determined from accompanying brightfield images (not shown). Yellow lines indicate x–y plane of z-section. Corresponding z-sections are shown below, minus DAPI stain. Bars (G-J), 7.5 μm; z-stack bars, 1 μm. Asterisks in panels G-J denote apical side of z-section.
PMP infiltration in solid tumor allografts and RNA transfer in mice. (A) Immunohistochemistry (IHC) in a 5-μm section from LLC tumor allograft, 21 days. αIIb integrin, red. (B) IHC in a 5-μm section from LLC tumor allograft, 21 days. αIIb integrin, red (top); CD63, green (middle); DAPI, blue (shown in merged image, bottom). αIIb integrin/CD63 overlap appears in some areas in the merged image as yellow. (C) Tumor cells from a resected LLC tumor at 21 days, isolated as described in supplemental Methods. αIIb integrin, red; DAPI, blue. (D-E) Tumor cells from resected LLC allograft 24 hours after transfusion of AO-labeled platelets. (D) αIIb integrin, red; (E) AO, green. Bars (A-E), 10 μm. *PMP− cells. (F) Percentage of cells with cytosolic AO staining, shown ± SEM (n = 3, >100 cells each). (G) Human platelets were transfected with unlabeled siRNA and transfused into mice bearing 20-day LLC tumors. After 24 hours, tumors were resected, digested, cleared of vascular cells with α-CD31 beads, and tumor cells were captured on fibronectin-coated coverslips. (H) Tumor cells ex vivo as in panel G, from mice transfused with human platelets transfected with FAM-siRNA. (I) LLC cells, treated in vitro with PMPs derived from human platelets 48 hours after platelet transfection with unlabeled siRNA. (J) Cells treated as in panel I with PMPs derived from human platelets transfected with FAM-siRNA. (H-J) Human αIIb integrin, red; FAM, green; DAPI, blue. Dashed white lines indicate cell borders as determined from accompanying brightfield images (not shown). Yellow lines indicate x–y plane of z-section. Corresponding z-sections are shown below, minus DAPI stain. Bars (G-J), 7.5 μm; z-stack bars, 1 μm. Asterisks in panels G-J denote apical side of z-section.
PMPs transfer platelet RNA, including miRNAs, to tumor cells in solid tumors in vivo and in vitro
We considered whether infiltrating PMPs could deliver RNA cargo to TCs. We transfused platelets, derived from wild-type (WT) mice and labeled with acridine orange (AO) DNA/RNA vital dye, into LLC tumor-bearing mice. PMP-associated TCs from resected tumors showed AO cytosolic fluorescence, indicating the presence of platelet-derived RNA in the PMP-targeted cell cytosol. TCs with no PMPs showed only background fluorescence (Figure 2D-F).
In addition to DNA/RNA, AO can also label platelet granules.63 To confirm that transferred platelet-derived material included RNA, and to investigate platelet miRNAs, we transfected human platelets with fluorophore 6-carboxyfluorescein (FAM)–labeled or unlabeled short inhibitory RNA (siRNA) as an miRNA mimic64 (supplemental Figure 2A-B) and transfused transfected platelets into tumor-bearing mice. Ex vivo TCs, which harbored PMPs, showed cytosolic FAM+ fluorescence in cells from FAM+-siRNA-transfused mice, but we did not observe FAM+ cells from control mice (Figure 2G-H).
To study PMP RNA transfer further, we used human platelets, from which we could more easily derive substantial quantities of PMPs. We stimulated ex vivo human platelets to release PMPs (supplemental Figure 3A), and incubated collected PMPs with LLCs in culture.23 After 1 hour of PMP exposure, most LLC cells were decorated with αIIb integrin puncta which resembled the intratumoral PMPs, and these puncta were removed by trypsin, suggesting a protein-mediated anchorage to the target cell surface (supplemental Figure 3B). Similar to TCs ex vivo, LLC cells exposed to PMPs from control-transfected platelets showed no FAM fluorescence (Figure 2I), whereas LLC cells exposed to PMPs from platelets transfected with FAM-siRNA showed cytosolic FAM+ fluorescence distinct from the αIIb integrin structures, selectively in PMP-targeted cells (Figure 2J). Thus, PMPs can deliver miRNA to TCs in vitro, consistent with previous reports in other systems.23,24 Together, these data demonstrate that platelet-derived siRNA (miRNA mimic) transfers horizontally to tumor cells in solid tumors via infiltrating PMPs, and transferred siRNA is not sequestered in PMPs but is distributed in the target cell cytosol.
Platelet-derived miRNAs transferred to tumor cells in solid tumors
We sought to characterize platelet-derived miRNAs transferred to TCs via MPs. Following in vitro PMP exposure, we extracted RNA from PMP-stripped TCs. PCR for previously identified abundant miRNAs in PMPs, many of which are involved in tumor progression,17,19,21,65-67 indicated enrichment of several miRNAs in LLCs following PMP exposure (Figure 3A). We generated LLC cells stably expressing E2-Crimson fluorophore to allow ex vivo TC isolation from resected tumors by single-cell sorting. Several miRNAs were upregulated in TCs in vivo compared with cells in culture as indicated by conventional PCR (Figure 3B) and quantitative reverse transcription PCR (qRT-PCR [qPCR]; Figure 3C), including miR-27a, miR-24, miR-25, miR-191, miR-9, and let-7a. Let-7a has been reported to constitute as much as 48% of the total platelet miRNA content.46 The moderate fold increase in let-7a and other miRNAs in TCs may reflect high endogenous expression levels of these miRNAs. However, miR-24 and miR-27a were evident only in PMP-treated LLC cells but not untreated cells in vitro and in ex vivo TCs by conventional PCR (Figure 3A-B). These miRNAs were each elevated more than ninefold ex vivo compared with cells in culture (Figure 3C), indicating that these miRNAs had been either expressed selectively in tumors or transferred to TCs in vivo.
PMP transfer of platelet miRNAs to tumor cells in solid tumors. (A) Total RNA extracted from LLC cells posttrypsinization, untreated (−) or exposed to PMPs for 16 hours (+), was subjected to poly(dA) tailing, cDNA synthesis, and PCR with the indicated miRNAs as 5′ forward primers, and poly(dT) universal 3′ reverse primers. (B) Total RNA extracted from tumor cells isolated from resected LLC tumors, or from LLCs maintained in culture, was subjected to poly(dA) tailing, cDNA synthesis, and PCR with the indicated miRNAs as 5′ forward primers, and poly(dT) universal 3′ reverse primers. −, LLCs maintained in culture; +, LLCs ex vivo from resected tumors. The red boxes indicate undetectable levels of miR-27a and miR-24 in LLC cells maintained in culture, compared with a band corresponding to each miRNA from LLC cells treated with PMPs (A) or from resected tumors (B). “no cDNA” samples used miR-24 oligonucleotides. (C) qRT-PCR using 5′ forward primers matching indicated miRNAs paired with poly(dT) universal 3′ reverse primers on cDNA from poly(A)-tailed RNA from LLC cells isolated from resected tumors, fold change over expression in LLCs in culture, shown ± SEM. miRNA primers were for 5p arms unless otherwise indicated. P < .05 for each (n = 4). Red line denotes parity. (D) Pf4-Cre/Uprt mice and 4TU RNA labeling, biotinylation, and isolation. (1) CA>GFPstop>Uprt mice and Pf4-Cre mice crossing to generate CA>Uprt/Pf4-Cre heterozygotes, which express UPRT selectively in megakaryocytes (MKs) (> and blue triangle, loxP site). (2) Tumor seeding in the het mice and (3) 4TU (U′) injection for selective incorporation in MK RNA. (4) 4TU-RNA transfers from the MK platelet progeny to tumors via PMPs. (5) Tumor resection and tumor cell isolation by fluorescence-activated cell sorting (FACS), followed by RNA extraction. (6) Platelet-derived 4TU-RNA labeling with N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP-biotin) added to the total tumor cell RNA, and isolation by affinity chromatography with avidin beads for further analysis. (E) PCR using miR-24 or miR-223 forward and poly(dT) reverse primers on avidin bead eluates from biotinylated RNA from tumor cells from 21-day tumors in 4TU-treated Pf4-Cre+/− (Pf4-Cre) and CA>HA-Uprt+/−Pf4-Cre+/− (Pf4-Cre/Uprt) mice. (F) qRT-PCR from panel E, showing fold change ± SEM in tumor cells extracted from Pf4-Cre/Uprt vs Pf4-Cre mice. Red line denotes parity. Let-7a fold change = 1.2 ± 0.02. P < .03 for each (n = 8). β-ME, β-mercaptoethanol.
PMP transfer of platelet miRNAs to tumor cells in solid tumors. (A) Total RNA extracted from LLC cells posttrypsinization, untreated (−) or exposed to PMPs for 16 hours (+), was subjected to poly(dA) tailing, cDNA synthesis, and PCR with the indicated miRNAs as 5′ forward primers, and poly(dT) universal 3′ reverse primers. (B) Total RNA extracted from tumor cells isolated from resected LLC tumors, or from LLCs maintained in culture, was subjected to poly(dA) tailing, cDNA synthesis, and PCR with the indicated miRNAs as 5′ forward primers, and poly(dT) universal 3′ reverse primers. −, LLCs maintained in culture; +, LLCs ex vivo from resected tumors. The red boxes indicate undetectable levels of miR-27a and miR-24 in LLC cells maintained in culture, compared with a band corresponding to each miRNA from LLC cells treated with PMPs (A) or from resected tumors (B). “no cDNA” samples used miR-24 oligonucleotides. (C) qRT-PCR using 5′ forward primers matching indicated miRNAs paired with poly(dT) universal 3′ reverse primers on cDNA from poly(A)-tailed RNA from LLC cells isolated from resected tumors, fold change over expression in LLCs in culture, shown ± SEM. miRNA primers were for 5p arms unless otherwise indicated. P < .05 for each (n = 4). Red line denotes parity. (D) Pf4-Cre/Uprt mice and 4TU RNA labeling, biotinylation, and isolation. (1) CA>GFPstop>Uprt mice and Pf4-Cre mice crossing to generate CA>Uprt/Pf4-Cre heterozygotes, which express UPRT selectively in megakaryocytes (MKs) (> and blue triangle, loxP site). (2) Tumor seeding in the het mice and (3) 4TU (U′) injection for selective incorporation in MK RNA. (4) 4TU-RNA transfers from the MK platelet progeny to tumors via PMPs. (5) Tumor resection and tumor cell isolation by fluorescence-activated cell sorting (FACS), followed by RNA extraction. (6) Platelet-derived 4TU-RNA labeling with N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP-biotin) added to the total tumor cell RNA, and isolation by affinity chromatography with avidin beads for further analysis. (E) PCR using miR-24 or miR-223 forward and poly(dT) reverse primers on avidin bead eluates from biotinylated RNA from tumor cells from 21-day tumors in 4TU-treated Pf4-Cre+/− (Pf4-Cre) and CA>HA-Uprt+/−Pf4-Cre+/− (Pf4-Cre/Uprt) mice. (F) qRT-PCR from panel E, showing fold change ± SEM in tumor cells extracted from Pf4-Cre/Uprt vs Pf4-Cre mice. Red line denotes parity. Let-7a fold change = 1.2 ± 0.02. P < .03 for each (n = 8). β-ME, β-mercaptoethanol.
To determine whether the additional miRNAs in TCs ex vivo were platelet-derived, we crossed CA-loxP (> )-GFPstop-loxP(> )-Uprt C57Bl/6 mice with Pf4-Cre C57Bl/6 mice to generate CA>Uprt/Pf4-Cre double heterozygotes for Cre-induced expression of Toxoplasma gondii uracil phosphoribosyltransferase (UPRT) selectively in megakaryocytes/platelets (Figure 3D). This enzyme is required for incorporation of 4TU, a cell-permeable, nonnative uracil variant, into nascent mammalian RNA; thus, only megakaryocytes can produce thio-RNA in these mice upon 4TU exposure, yielding platelets selectively harboring thio-RNA which can be isolated by thiol-biotinylation and avidin chromatography (Figure 3D).68 We seeded LLC-E2-Crimson cell tumors in the flanks of CA>Uprt/Pf4-Cre heterozygotes or Pf4-Cre controls, followed by 4TU i.p. injection, TC collection, and biotin/avidin isolation of platelet-derived RNA in PMP-stripped TCs (Figure 3D). Both miR-24 and miR-223 were observed as thio-RNA extracted from isolated TC RNA from Pf4-Cre/CA>Uprt mice, but not from TCs from Pf4-Cre mice, demonstrating in vivo transfer of these platelet-derived miRNAs to TCs (Figure 3E). qPCR from thio-RNA samples revealed significant increases over control in each of the screened miRNAs, with the greatest increase observed in miR-24 (Figure 3F). 4TU-RNA in TCs was platelet-derived and not from PMP-mediated transfer of the UPRT enzyme from platelets to tumor cells, as hemagglutinin (HA)-UPRT was expressed selectively in platelets in Pf4-Cre/CA>Uprt mice, but we did not detect HA-UPRT in PMPs nor in ex vivo TCs in these mice or in control mice (supplemental Figure 4); thus, UPRT-driven RNA labeling was restricted to megakaryocytes/platelets in this system. Together, these data demonstrate that platelet-derived miRNAs transfer to TCs in solid tumors in vivo, and miR-24 was a major species in the transfer. Attachment of platelet-derived miRNAs to TC surfaces could not be ruled out in these experiments; however, TCs can internalize platelet-derived miRNAs in vivo (Figure 2).
PMPs induce tumor cell apoptosis in an miR-24–dependent manner
We considered what effects PMPs may have on TC proliferation. PMP exposure in vitro blunted proliferation in a dose-dependent manner in LLCs (Figure 4A), as well as MC-38 colon carcinoma cells (Figure 4B), indicating broad tumor type specificity in PMP-induced TC growth suppression. Growth inhibition by PMPs was reversed by transfection with antagomiR-2469 (Figure 4C-D), indicating that miR-24 is a major driver of growth inhibition by PMP exposure in these carcinoma cells. We observed a time lag in growth inhibition (Figure 4C-D), which may be due in part to the time required for miRNA downregulation of target RNAs and degradation of residual proteins. Mir-24–dependent TC growth inhibition correlated with expression of cleaved caspase-3, a marker for apoptosis, in both TC lines (Figure 4E-F), whereas we did not observe significant effects of PMPs on cell cycle progression (supplemental Figure 5), indicating that the reduction in cell number by PMP exposure was associated with induction of TC apoptosis. Association of PMPs with caspase-3 and apoptosis more broadly has been observed in other systems and various disease states.70,71
PMPs induce tumor cell apoptosis and inhibit tumor growth via miR-24. (A) The indicated number of PMPs collected from freshly isolated human platelets was coincubated with 5 × 104 LLC cells every 24 hours. Cells were counted at 60 hours (n = 8). (B) MC-38 cells, treated as assessed as in panel A (n = 3). (A-B) *P < .0007; **P < .001; ***P < .05; #P < .02; ##P < .01. (C) A total of 5 × 104 LLC cells were transfected and treated every 24 hours with 109 freshly isolated PMPs as indicated, and counted daily (n = 5). (D) MC-38 cells transfected, treated, and analyzed as in panel C. (C-D): *P < .03; **P < .04 (n = 3). (E) LLC cells treated as in panel C were harvested and lysates were processed for western blotting with antibodies to cleaved caspase-3 (cl. cas-3) and β-actin. (F) Lysates of MC-38 cells treated as in panel D were processed for western blotting as in panel E (n = 5) for panels E-F. (G) A total of 1 × 106 LLCs were transfected as indicated, and after 18 hours were seeded as allografts by bolus injection into the flanks of WT mice. Beginning at day 8, 1 × 1010 PMPs freshly isolated from human platelets were counted and transfused daily by tail vein injection. Tumor volumes were measured daily with calipers (n = 6). *P < .02. (H) MC-38 cells were transfected and implanted, followed by PMP transfusion, and tumor growth was monitored as in panel G. **P < .003 (n = 6). All plots, shown ± SEM. n.s., not significant.
PMPs induce tumor cell apoptosis and inhibit tumor growth via miR-24. (A) The indicated number of PMPs collected from freshly isolated human platelets was coincubated with 5 × 104 LLC cells every 24 hours. Cells were counted at 60 hours (n = 8). (B) MC-38 cells, treated as assessed as in panel A (n = 3). (A-B) *P < .0007; **P < .001; ***P < .05; #P < .02; ##P < .01. (C) A total of 5 × 104 LLC cells were transfected and treated every 24 hours with 109 freshly isolated PMPs as indicated, and counted daily (n = 5). (D) MC-38 cells transfected, treated, and analyzed as in panel C. (C-D): *P < .03; **P < .04 (n = 3). (E) LLC cells treated as in panel C were harvested and lysates were processed for western blotting with antibodies to cleaved caspase-3 (cl. cas-3) and β-actin. (F) Lysates of MC-38 cells treated as in panel D were processed for western blotting as in panel E (n = 5) for panels E-F. (G) A total of 1 × 106 LLCs were transfected as indicated, and after 18 hours were seeded as allografts by bolus injection into the flanks of WT mice. Beginning at day 8, 1 × 1010 PMPs freshly isolated from human platelets were counted and transfused daily by tail vein injection. Tumor volumes were measured daily with calipers (n = 6). *P < .02. (H) MC-38 cells were transfected and implanted, followed by PMP transfusion, and tumor growth was monitored as in panel G. **P < .003 (n = 6). All plots, shown ± SEM. n.s., not significant.
PMPs inhibit tumor growth in an miR-24–dependent manner
We tested tumor growth effects of PMPs in vivo. Daily transfusion of freshly generated human PMPs into mice bearing LLC or MC-38 tumors, beginning at day 8, markedly inhibited tumor growth, which was abrogated by transfection with antagomiR-24 prior to allograft implantation (Figure 4E-F). These results indicate that miR-24 in the transfused PMPs was partially responsible for tumor growth inhibition, which we would predict as miR-24 is conserved between mouse and human, and therefore these homologs have identical seed target sequences and thus, with identical binding affinities, putatively can target the same RNAs. Transfused PMPs infiltrated tumors and appeared broadly distributed in the intratumoral environment (supplemental Figure 6A), but neither endogenous nor exogenous PMPs were detected in nontumor tissue (supplemental Figure 6B). Thus, PMPs are tumor suppressive both in vitro and in vivo via effects of transferred miRNAs including miR-24.
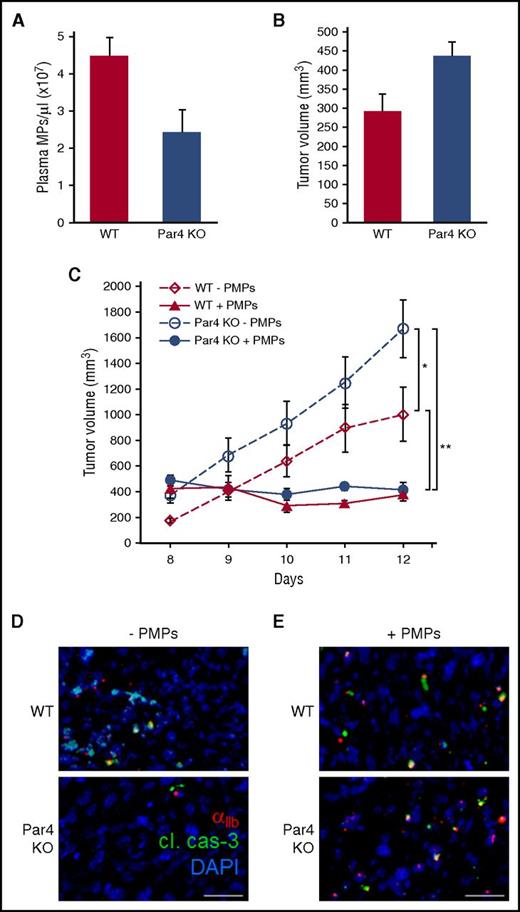
PMP depletion in Par4-null mice prevents PMP-mediated tumor growth inhibition in vivo via induction of apoptosis in PMP-targeted tumor cells
To investigate further the specific role of PMPs in tumor growth inhibition, we evaluated MPs and tumor growth in mice deleted for the Par4 thrombin receptor, which has been shown to be the major driver of PMP generation.72 Plasma MPs were reduced by ∼50% in Par4 knockout (KO) mice compared with WT (Figure 5A), whereas total circulating platelet counts were unaltered (results not shown). In accordance with an inhibitory role for PMPs, solid tumor growth was accelerated in Par4 KO mice compared with WT (Figure 5B-C). Daily transfusion of PMPs was sufficient to halt tumor growth in both WT and Par4 KO mice (Figure 5C), indicating that PMPs are the principal effectors of tumor growth inhibition in this model. Tumor infiltration of endogenous PMPs was reduced in Par4 KO mice compared with WT, consistent with depletion of PMPs in those mice (Figure 5D), and PMP transfusion restored PMP infiltration in tumors (Figure 5E). PMP-associated TCs showed expression of cleaved caspase-3, whereas adjacent, nontargeted cells did not (Figure 5D-E); thus, PMP targeting is associated with TC apoptosis in vivo.
Plasma MPs and tumor growth in Par4 KO mice. (A) Plasma MPs from WT and Par4 KO mice were analyzed by nanoparticle tracking, and are shown ± SEM (n = 6); P < .03. (B) A total of 1 × 106 LLC cells were seeded in the flanks of WT and Par4 KO mice, and tumor volumes were measured 8 days after seeding, shown ± SEM (n = 6); P < .03. (C) LLC tumors were seeded as in panel B, and 1 × 1010 freshly isolated PMPs were transfused in the tail vein every 24 hours beginning at day 8 as indicated. Tumor volumes were measured daily, and are shown ± SEM (n = 6); *P < .03; **P < .002. (D) Tumors from panel C were resected, fixed, and processed for IHC with antibodies to murine CD41 (αIIb integrin, red) to label endogenous PMPs, and cleaved caspase-3 (green); DAPI stain is shown in blue. (E) Tumor sections from panel C stained with antibodies to human CD41 (αIIb integrin, red) to label transfused PMPs, cleaved caspase-3 (cl. cas-3, green), and DAPI (blue). Bars, 15 μm.
Plasma MPs and tumor growth in Par4 KO mice. (A) Plasma MPs from WT and Par4 KO mice were analyzed by nanoparticle tracking, and are shown ± SEM (n = 6); P < .03. (B) A total of 1 × 106 LLC cells were seeded in the flanks of WT and Par4 KO mice, and tumor volumes were measured 8 days after seeding, shown ± SEM (n = 6); P < .03. (C) LLC tumors were seeded as in panel B, and 1 × 1010 freshly isolated PMPs were transfused in the tail vein every 24 hours beginning at day 8 as indicated. Tumor volumes were measured daily, and are shown ± SEM (n = 6); *P < .03; **P < .002. (D) Tumors from panel C were resected, fixed, and processed for IHC with antibodies to murine CD41 (αIIb integrin, red) to label endogenous PMPs, and cleaved caspase-3 (green); DAPI stain is shown in blue. (E) Tumor sections from panel C stained with antibodies to human CD41 (αIIb integrin, red) to label transfused PMPs, cleaved caspase-3 (cl. cas-3, green), and DAPI (blue). Bars, 15 μm.
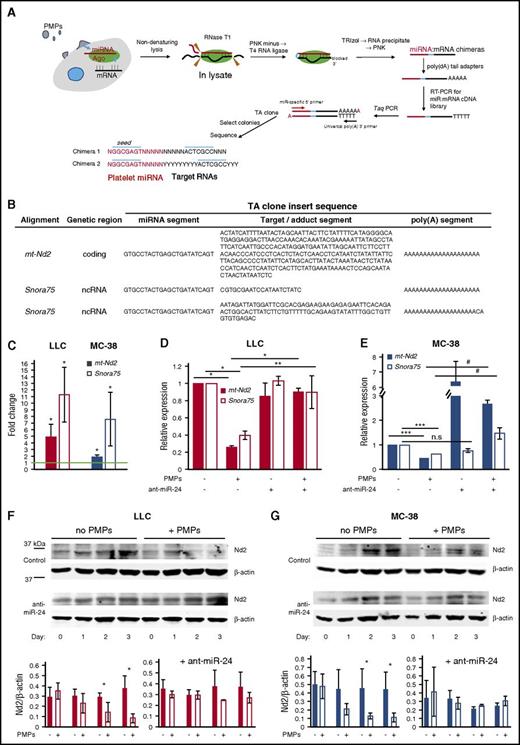
Identification of tumor cell RNA targets of PMP-derived miR-24
To identify TC RNA targets of PMP-derived miR-24, we modified a method recently described,48 in which single-strand chimeras derived from truncated miRNA:target RNA hybrid pairs are generated in cell lysate fractions (Figure 6A). We focused on miR-24 targets, as miR-24 was a major species transferred from PMPs and a major contributor to growth inhibitory effects of PMPs. Human and murine miR-24-1-5p and -2-5p sequences are identical. We performed miRNA:mRNA ligation reactions on lysates from LLC cells with or without PMP exposure, followed by PCR with miR-24 5′ and universal 3′ primers, and direct cloning and sequencing of the PCR products. Whereas some insert sequences were either too short or contained misalignments, preventing unambiguous identification of the miRNA-ligated target RNAs (supplemental Table 1), several of the miR-24 adduct inserts could be unambiguously assigned. A 21-nt insert corresponded exclusively to Snora75, an H/ACA box small nucleolar RNA (snoRNA), and another independent clone contained a 96-nt segment also corresponding exclusively to Snora75. Hence, some miR-24 adducts included a noncoding RNA (ncRNA). A 281-nt insert sequence in another clone corresponded exclusively and entirely to a segment of mitochondrial mt-Nd2 mRNA, suggesting targeting of the coding region of a mitochondrial gene by miR-24 (Figure 6B; supplemental Figure 7).73
RNA targets of PMP-derived miR-24. (A) Schematic for low-throughput miRNA target identification. PMP-treated cells were lysed posttrypsinization by suspension in hyposmotic buffer followed by sonication. Whole-cell extracts were treated with RNase T1, 3′ end blocking with PNK minus lacking 3′ phosphatase activity, followed by T4 RNA ligase. RNA extracted from these samples with TRIzol was tagged with poly(dA) tails and subjected to first-strand cDNA synthesis, followed by conventional PCR with Taq polymerase using miRNA-specific 5′ primers and poly(dT) 3′ primer, direct cloning of unsorted PCR products into the pCR2.1 (TA) vector, and transformation of the DNA ligation reactions into E coli. Colonies were selected and plasmid DNA preparations were analyzed by conventional sequencing. (B) miR-24:target RNA adduct clones. TA clones with inserts matching unique sequences are shown. The insert sequences are separated in the table into the apparent miRNA segment, the cognate target/adduct segment, and the poly(A) segment. (C) mt-Nd2 and Snora75 RNA enrichment in RISC complexes following tumor cell exposure to PMPs. Shown are qPCR ratios for mt-Nd2 and Snora75 RNA content in Ago2 immunoprecipitate (IP) fractions from LLC (left) or MC-38 cells (right) treated with PMPs, relative to IP fractions from untreated cells. *P < .05 (n = 3). (D) LLC cells were transfected with 25 μg of phosphorothioate, LNA 8-nt control or antagomiR-24 (ant-miR-24), 24 hours prior to PMP exposure. RNA was isolated from cells 16 hours after exposure to PMPs or blank media. qRT-PCR was performed using 100-bp PCR fragments of each transcript, and relative expression levels were quantified using GAPDH as a housekeeping gene control, normalized to target RNA expression in untreated cells, shown as 1. *P < .001; **P < .05 (n = 7). (E) MC-38 cells treated and analyzed as in panel D. ***P < .01; #P < .02 (n = 5). All histograms, shown ± SEM. (F-G) Western blotting with α-mt-Nd2 antibodies (Nd2) of lysates of cells treated with PMPs for up to 3 days. β-actin was used as a loading control. Densitometry results are shown for Nd2/β-actin ratios, ± SEM for 3 independent experiments. *P < .05, all others n.s.
RNA targets of PMP-derived miR-24. (A) Schematic for low-throughput miRNA target identification. PMP-treated cells were lysed posttrypsinization by suspension in hyposmotic buffer followed by sonication. Whole-cell extracts were treated with RNase T1, 3′ end blocking with PNK minus lacking 3′ phosphatase activity, followed by T4 RNA ligase. RNA extracted from these samples with TRIzol was tagged with poly(dA) tails and subjected to first-strand cDNA synthesis, followed by conventional PCR with Taq polymerase using miRNA-specific 5′ primers and poly(dT) 3′ primer, direct cloning of unsorted PCR products into the pCR2.1 (TA) vector, and transformation of the DNA ligation reactions into E coli. Colonies were selected and plasmid DNA preparations were analyzed by conventional sequencing. (B) miR-24:target RNA adduct clones. TA clones with inserts matching unique sequences are shown. The insert sequences are separated in the table into the apparent miRNA segment, the cognate target/adduct segment, and the poly(A) segment. (C) mt-Nd2 and Snora75 RNA enrichment in RISC complexes following tumor cell exposure to PMPs. Shown are qPCR ratios for mt-Nd2 and Snora75 RNA content in Ago2 immunoprecipitate (IP) fractions from LLC (left) or MC-38 cells (right) treated with PMPs, relative to IP fractions from untreated cells. *P < .05 (n = 3). (D) LLC cells were transfected with 25 μg of phosphorothioate, LNA 8-nt control or antagomiR-24 (ant-miR-24), 24 hours prior to PMP exposure. RNA was isolated from cells 16 hours after exposure to PMPs or blank media. qRT-PCR was performed using 100-bp PCR fragments of each transcript, and relative expression levels were quantified using GAPDH as a housekeeping gene control, normalized to target RNA expression in untreated cells, shown as 1. *P < .001; **P < .05 (n = 7). (E) MC-38 cells treated and analyzed as in panel D. ***P < .01; #P < .02 (n = 5). All histograms, shown ± SEM. (F-G) Western blotting with α-mt-Nd2 antibodies (Nd2) of lysates of cells treated with PMPs for up to 3 days. β-actin was used as a loading control. Densitometry results are shown for Nd2/β-actin ratios, ± SEM for 3 independent experiments. *P < .05, all others n.s.
To evaluate mt-Nd2 mRNA and Snora75 regulation by miRNAs in RNA-induced silencing complexes (RISCs) in PMP-exposed cells, we extracted RNA from Ago2 immunoprecipitate (IP) fractions in untreated and PMP-treated TCs, and analyzed relative enrichment by qPCR. As before, we used glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a housekeeping control, which is valid in this case as GAPDH is recruited into RISCs and modulated by siRNAs or miRNAs, but known miRNAs targeting GAPDH have not been detected in platelets or PMPs.64,74-76 PMP exposure led to increases in mt-Nd2 and Snora75 RNAs in Ago2-containing complexes compared with untreated cells (Figure 6C), indicating enhanced recruitment of these RNA targets to RISCs by PMP exposure. These results were not due to transfer of preformed miRNA:target RNA complexes via PMPs, as evidenced by lack of human mt-Nd2 and Snora75 RNAs in induced PMPs as well as in human PMP-treated TCs. In contrast, miR-24 was evident in platelets, PMPs, and PMP-treated TCs, further supporting direct transfer of this miRNA from platelets to TCs via PMPs (supplemental Figure 8).
RNA levels of Snora75 and mt-Nd2 were substantially suppressed in both LLC and MC-38 cells following PMP exposure (Figure 6D-E), indicating broad tumor type specificity in PMP-mediated gene suppression. AntagomiR-24 rescued mt-Nd2 and Snora75 downregulation by PMP exposure (Figure 6D-E). Interestingly, antagomiR-24 had no effect on expression of these RNAs in untreated LLCs, but led to mt-Nd2 upregulation in MC-38 cells both with and without PMPs (Figure 6D-E). These results suggest that mt-Nd2 steady-state expression is modulated by endogenous miR-24 in MC-38 cells to a greater extent than in LLC cells, and addition of exogenous PMP-derived miR-24 enhances gene suppression in both cell lines. Mt-Nd2 protein expression was also decreased by PMPs in both TC lines, and these decreases were prevented by antagomiR-24 (Figure 6F-G). Thus, PMP exposure in these TCs induces downregulation of mt-Nd2 and Snora75 via miR-24.
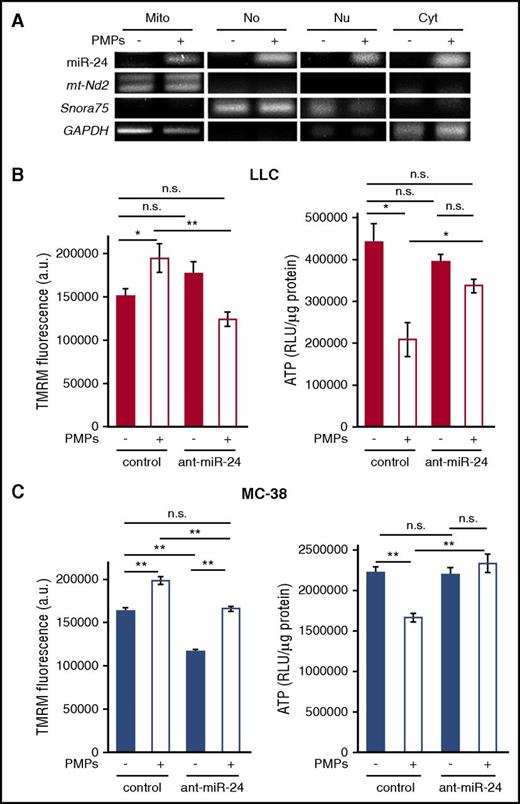
PMP-derived miR-24 localizes to mitochondria and inhibits tumor cell mitochondrial function
MiR-24 was broadly distributed in multiple compartments in PMP-exposed cells, including nucleus, nucleoli, mitochondria, and mitochondria-depleted cytoplasm (Figure 7A). MiR-24 was recently observed in nucleoli77 ; these data indicate that miR-24 is also a mitomiR.78 This multiorganelle localization is consistent with the ability of miR-24 to target and downregulate RNAs in each compartment.
PMP-transferred miR-24 inhibits mitochondrial function in tumor cells. (A) cDNA from RNA isolated from mitochondria (Mito), nucleolar (No), nuclear (Nu), and postmitochondria (Cyto) fractions of untreated and PMP-treated LLC cells (−/+) was subject to PCR for the indicated genes. (B) Mitochondrial membrane potential (TMRM, left) and ATP levels (right) were assessed in LLC cells ± PMPs and antagomiR-24 as indicated. (C) TMRM (left) and ATP (right) in MC-38 cells, treated as in panel B. *P < .01; **P < .0001 (n = 3). All histograms, shown ± SEM. RLU, relative luminescence unit.
PMP-transferred miR-24 inhibits mitochondrial function in tumor cells. (A) cDNA from RNA isolated from mitochondria (Mito), nucleolar (No), nuclear (Nu), and postmitochondria (Cyto) fractions of untreated and PMP-treated LLC cells (−/+) was subject to PCR for the indicated genes. (B) Mitochondrial membrane potential (TMRM, left) and ATP levels (right) were assessed in LLC cells ± PMPs and antagomiR-24 as indicated. (C) TMRM (left) and ATP (right) in MC-38 cells, treated as in panel B. *P < .01; **P < .0001 (n = 3). All histograms, shown ± SEM. RLU, relative luminescence unit.
Mt-Nd2 is a component of the nicotinamide adenine dinucleotide: ubiquinone oxidoreductase (complex I), responsible for mitochondrial oxidative phosphorylation and reactive oxygen species production.79 We measured mitochondrial membrane potential in PMP-treated and untreated cells. Nuclear localization of the membrane potential indicator, tetramethylrhodamine, methyl ester (TMRM), was significantly increased by PMP treatment (Figure 7B-C left panels), indicating mitochondrial depolarization following PMP exposure. This could be due to inactivation of complex I and shunt to complex III.80 In support of this, ATP generation was strongly inhibited by PMP treatment (Figure 7B-C right panels). AntagomiR-24 abrogated these effects of PMP exposure (Figure 7B-C). AntagomiR-24 also led to mitochondrial depolarization in the absence of PMPs in MC-38 cells (Figure 7C), correlating with moderate mt-Nd2 suppression by PMPs in these cells, and increased mt-Nd2 by antagomiR-24 alone (Figure 6). However, ATP production was not increased over baseline levels in this case. Platelet-derived mitochondria were not transferred via PMPs to TCs, as we did not detect Mitotracker Red–labeled platelet mitochondria in TCs after PMP generation and exposure (supplemental Figure 9), and as noted previously, human mt-Nd2 RNA, which is contained within mitochondria, was also not transferred from platelets to PMPs or TCs (supplemental Figure 8). Together, these data demonstrate that miR-24, transferred from PMPs to tumor cells, causes mitochondrial dysfunction and tumor growth inhibition.
Discussion
Our findings establish a previously unappreciated mode of indirect intercellular communication between platelets and tumor cells in solid tumors, with inhibitory effects on tumor progression by transfer of platelet miRNAs and downregulation of tumor cell gene expression. PMP infiltration, transfer of platelet-derived miRNAs to TCs, and gene-regulatory and growth-suppressive effects, together extend the reach and capabilities of platelets to affect cancer progression beyond the intravascular space. Our findings further support broad specificities in miRNA targeting, including mRNA coding regions (eg, mitochondrial mRNAs) and other ncRNAs as viable targets for effective suppression. Our findings also implicate PMPs and their cargo more broadly, as important contributors to functional outcomes in pathological conditions of increased vascular permeability.
We observed PMP infiltration in multiple solid tumor types, and at each tumor grade, but not in unaffected normal tissues. This difference likely reflects increased permeability of tumor neovasculature, which unlike mature blood vessels, is perforated with pores >>100 nm as a result of dysfunctional endothelial junctions, allowing for leak of blood-borne protein complexes and other macromolecular complexes into the intratumoral space.50 This pore size is permissive for extravascular leak of PMPs, and it is likely that other cell-derived MPs as well as exosomes (30-100 nm diameter) invade the tumor microenvironment through these pores. PMP exposure due to vascular leak is therefore likely restricted to solid tumors, distinct from normal tissues, adding PMPs and other microvesicles to the unique composition of the tumor microenvironment. Mechanisms of PMP attachment and internalization in tumor cells remain to be elucidated, but could involve interactions with multiple receptors such as glycoprotein 1b (potentially in complex with VWF),81 P-selectin,82 phosphatidylserine receptors on the tumor cell surface,83 or a combination of these and other interactions.
Interactions between MPs and cells have been investigated in many contexts, these interactions yield a broad range of outcomes.84,85 Endothelium and blood cells are exposed to PMPs, and effects are just beginning to be explored.19,86 PMPs have been shown to transfer miRNAs to cells when coincubated in vitro, and to modulate target cell gene expression.23,24 For example, PMP incubation with A549 lung carcinoma cells in culture enhanced invasiveness due to increased miR-223 and suppression of anti-invasive genes.24 The present study expands on this background by demonstrating that PMP-cell interactions have functionally important roles in vivo, via miRNA transfer. Differences in the degree of miRNA increases ex vivo compared with in vitro may reflect upregulation by TCs during tumor progression, and/or internalization of exogenous miRNA from stromal sources other than PMPs. However, our results point to miR-24 as a major PMP-derived regulator of tumor growth in two cancer cell lines. Together, our data support growth inhibitory roles of PMP interactions with TCs in solid tumors, via direct transfer of platelet-derived miRNAs and modulation of TC gene expression, resulting in tumor cell apoptosis, thus representing a new inhibitory function of this interaction in tumor progression.
Targets of transferred miR-24 included noncanonical RNAs, a mitochondrial mRNA lacking a 3′-untranslated region (UTR), and a small nucleolar ncRNA (snoRNA), representing regulation of one ncRNA by another. MiRNAs can regulate many RNA classes, and can bind many types of target sites, not limited to 3′-UTRs and with variations in seed complementarity.48,87-89 Whether noncanonical miRNA response elements are biologically relevant remains controversial, as recent studies indicate that noncanonical sites may not contribute substantially to gene suppression.90 However, the miR-24 targets we identified were found through an unbiased screen following direct ligation of miRNAs to their targets within RISCs. These RNAs were enriched in Ago2-containing complexes following PMP exposure, indicating recruitment to RISCs and enhanced RISC targeting by PMP-derived miRNAs. Together these findings suggest flexibility in functional seed recognition in these cells. Alternatively, PMPs, like platelets, may be enriched for variant miRNA isoforms (isomiRs) with base-shifted seed sites, including isomiRs of miR-24 and other miRNAs.46,73 Indeed, a 7-mer sequence in mt-Nd2 (AGTAGGC, conserved in human and murine) corresponds to a seed response element for a putative isomiR of miR-24.91 Nonetheless, the functional outcome of miRNA transfer from PMPs to TCs included suppression of multiple genes and inhibition of tumor growth.
The results of this study demonstrate new functions for platelets beyond the vascular space, and expanded roles in tumor progression, further complicating the platelet-cancer relationship. Platelets support cancer progression at several levels, particularly at late stages in primary tumors and in metastatic dissemination.4,92 Our results demonstrate tumor-suppressive roles at earlier stages, via growth-suppressive effects through downregulation of TC genes and induction of tumor cell apoptosis. Platelet miRNA transfer may also modulate other aspects of tumor biology, such as multidrug resistance, which is known to be regulated by MPs.93-96 The full cohort of platelet miRNAs is likely to modulate expression of multiple genes, potentially in a tumor type-specific manner, in TCs as well as in the tumor microenvironment. Thus, platelets contribute both positively and negatively to cancer progression through various modes and at multiple stages.
PMP extravasation may be a common consequence in the context of endothelial barrier dysfunction resulting in increased vascular permeability. Relationships of platelets and MPs with vascular leak have recently been described in cardiovascular disease,97 ischemia and postischemic tissue repair,98 sepsis,99-101 wound healing,102 and diabetes,103-105 implicating PMPs and miRNA transfer more broadly beyond solid tumor progression as a potential regulator of physiological responses to vascular leak. Such effects merit further exploration.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank John Kostyak, Maria de la Fuente, and Ilya Serebriiskii for technical assistance, Satya Kunapuli for support, and the Kunapuli laboratory for discussions. The authors thank Erica Golemis for critical reading of the manuscript.
This work was supported by grants from the National Institutes of Health (National Heart, Lung, and Blood Institute R01HL109568 [A.K.R.]; National Heart, Lung, and Blood Institute R01HL128446 [A.S.]; National Institute of General Medical Sciences R01GM109882, National Heart, Lung, and Blood Institute R01HL086699, National Heart, Lung, and Blood Institute R01HL119306, and National Institute of General Medical Sciences 1S10RR027327 [M.M.]; National Institute of General Medical Sciences K01GM103806 [J.W.R.]; and National Heart, Lung, and Blood Institute U54HL112311 [A.S.W.]) and from the American Heart Association (14GRNT20460004 [J.Y. and L.C.E.] and 16GRNT27260319 [L.E.G.]).
Authorship
Contribution: L.E.G. conceptualized the study; A.K.R., M.M., J.W.R., M.T.N., L.C.E., A.S.W., and L.E.G. designed the study methodology; J.V.M., J.G.T.W., G.F.M., N.E.H., S.R., D.T., J.W.R., J.Y., and L.E.G. investigated; J.V.M., J.G.T.W., and L.E.G. wrote the original draft of the manuscript; J.V.M., J.G.T.W., M.T.N., A.S., A.K.R., J.W.R., A.S.W., and L.E.G. reviewed and edited the manuscript; A.K.R., M.M., J.W.R., A.S.W., J.Y., L.C.E., and L.E.G. acquired funding; M.T.N., M.A.K., and A.S. provided resources; and A.K.R., M.M., A.S., L.C.E., A.S.W., and L.E.G. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lawrence E. Goldfinger, Department of Anatomy & Cell Biology and The Sol Sherry Thrombosis Research Center, Lewis Katz School of Medicine at Temple University, 3420 N Broad St, Philadelphia, PA 19140; e-mail: goldfinger@temple.edu.
References
Author notes
J.V.M. and J.G.T.W. contributed equally to this work.



![Figure 3. PMP transfer of platelet miRNAs to tumor cells in solid tumors. (A) Total RNA extracted from LLC cells posttrypsinization, untreated (−) or exposed to PMPs for 16 hours (+), was subjected to poly(dA) tailing, cDNA synthesis, and PCR with the indicated miRNAs as 5′ forward primers, and poly(dT) universal 3′ reverse primers. (B) Total RNA extracted from tumor cells isolated from resected LLC tumors, or from LLCs maintained in culture, was subjected to poly(dA) tailing, cDNA synthesis, and PCR with the indicated miRNAs as 5′ forward primers, and poly(dT) universal 3′ reverse primers. −, LLCs maintained in culture; +, LLCs ex vivo from resected tumors. The red boxes indicate undetectable levels of miR-27a and miR-24 in LLC cells maintained in culture, compared with a band corresponding to each miRNA from LLC cells treated with PMPs (A) or from resected tumors (B). “no cDNA” samples used miR-24 oligonucleotides. (C) qRT-PCR using 5′ forward primers matching indicated miRNAs paired with poly(dT) universal 3′ reverse primers on cDNA from poly(A)-tailed RNA from LLC cells isolated from resected tumors, fold change over expression in LLCs in culture, shown ± SEM. miRNA primers were for 5p arms unless otherwise indicated. P < .05 for each (n = 4). Red line denotes parity. (D) Pf4-Cre/Uprt mice and 4TU RNA labeling, biotinylation, and isolation. (1) CA>GFPstop>Uprt mice and Pf4-Cre mice crossing to generate CA>Uprt/Pf4-Cre heterozygotes, which express UPRT selectively in megakaryocytes (MKs) (> and blue triangle, loxP site). (2) Tumor seeding in the het mice and (3) 4TU (U′) injection for selective incorporation in MK RNA. (4) 4TU-RNA transfers from the MK platelet progeny to tumors via PMPs. (5) Tumor resection and tumor cell isolation by fluorescence-activated cell sorting (FACS), followed by RNA extraction. (6) Platelet-derived 4TU-RNA labeling with N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP-biotin) added to the total tumor cell RNA, and isolation by affinity chromatography with avidin beads for further analysis. (E) PCR using miR-24 or miR-223 forward and poly(dT) reverse primers on avidin bead eluates from biotinylated RNA from tumor cells from 21-day tumors in 4TU-treated Pf4-Cre+/− (Pf4-Cre) and CA>HA-Uprt+/−Pf4-Cre+/− (Pf4-Cre/Uprt) mice. (F) qRT-PCR from panel E, showing fold change ± SEM in tumor cells extracted from Pf4-Cre/Uprt vs Pf4-Cre mice. Red line denotes parity. Let-7a fold change = 1.2 ± 0.02. P < .03 for each (n = 8). β-ME, β-mercaptoethanol.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/5/10.1182_blood-2016-11-751099/4/m_blood751099f3.jpeg?Expires=1764956976&Signature=L7fziaz~VkEMG1tWSjywZxJKR-NadEFonmHsFJVeInfgBVJu2Mx~sdzqZGRc9ca4jkubX2iFd0aA2ybUsmQIQGgxyyTNG60YVNIIsjxjH1lOMrELLx1VT8Xqno~lK7X65JVaUlmg56HFVzDjOGuVe4j9gqfjFfxoEcMjQ11Uul4ukL-itW9DSJzh3eWlwJTxu~dDX-UCzV3Tl6sG1zgYMt0m9U6KZvImOjDtrjbW6ZVb5qzM11-Eabh-uV7YkWHjVpDxn-ulOH7ahhQX5dTJyVL6WV5lV7uIwV~1Ybab41JPl4XuKi3YcwIAO-fivfkNXhAQKvAWIbfyCqF1tyxwww__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal