To the editor:
Splenic diffuse red pulp lymphoma (SDRPL) is a rare small B-cell neoplasm provisionally included in the category of unclassifiable splenic B-cell lymphomas/leukemias in the 2008 World Health Organization classification.1 SDRPL is characterized by a diffuse pattern of involvement of the splenic red pulp by small monomorphous B lymphocytes. This indolent but incurable disease is more common in older men and usually manifests itself with splenomegaly and moderate lymphocytosis. Patients are normally diagnosed at stage IV when spleen, bone marrow, and peripheral blood are involved. The differential diagnosis from other splenic lymphomas, such as splenic marginal zone lymphoma (SMZL), hairy cell leukemia (HCL), and its variant (HCL-v), can be difficult because of their similar clinical presentations and the absence of specific molecular markers and may require examination of the spleen histology.2,3 Our knowledge of the molecular mechanisms involved in SDRPL is very limited because the incidence of this lymphoma is low and few splenectomy specimens are available. A better molecular characterization of this tumor could make their specific recognition easier, thereby enabling them to be differentiated from other splenic lymphomas, ultimately providing diagnostic markers and a basis for targeted therapy. To date, only 2 sequencing studies have been published in which NOTCH1, NOTCH2, TP53, MYD88, and MAP2K1 mutations were described.4,5
Here, we studied the mutational status of the complete exome of 23 SDRPL patients using the Illumina platform for next-generation whole-exome sequencing (WES). Details of the patients and the sequencing procedures are described in supplemental Table 1 and supplemental Methods (available on the Blood Web site). Statistics of next-generation sequencing data are shown in supplemental Table 2.
After filtering and visual inspection on the Integrative Genomics Viewer (www.broadinstitute.org/igv/), 455 substitutions and 39 insertion deletions from 464 genes were selected. Most of these mutations (77% on average) were validated in the cases where DNA was available (supplemental Table 3). Fifteen variants found in our series are described as somatic mutations in the Catalogue Of Somatic Mutations In Cancer (COSMIC) database (supplemental Table 3). No mutation was found in NOTCH2, a gene recurrently mutated in SMZL,6-10 or in BRAF V600E, a mutation characteristic of HCL.11,12
Figure 1A shows the most frequently mutated genes. Twenty-nine genes appear mutated in >1 patient, and 16 of these genes harbored recurrent mutations. ARID1A, MAP2K1, SYK, and CCND3 are recurrently mutated genes of special interest in lymphomas. CCND3 was mutated in 4 patients. Cyclin D3 forms a complex with, and functions as, a regulatory subunit of CDK4 or CDK6, whose activity is required for cell-cycle G1/S transition. More specifically, cyclin D3 has been found to play a key function in B-cell development by integrating cytokine and pre-B-cell receptor–dependent signals to expand the pool of pre-B cells that have successfully rearranged the immunoglobulin heavy chain.13 Moreover, CCND3 mutations have been previously described in Burkitt lymphoma, diffuse large B-cell lymphoma, HCL-v, and follicular lymphoma, as well as in various solid tumors.12,14-16
CCND3 mutations in SDRPL. (A) Genes recurrently mutated in SDRPL cases. From left to right are written all genes harboring mutations in 2 or more patients. Colors represent the main function of each gene, assigned on the bases of the information compiled in the GeneCards and KEGG databases. (B) Schematic representation of the mapping of CCND3 variants detected in SDRPL patients. (C) Details of each variant. BCR, B-cell receptor; cDNA, complementary DNA; CDS, coding sequence.
CCND3 mutations in SDRPL. (A) Genes recurrently mutated in SDRPL cases. From left to right are written all genes harboring mutations in 2 or more patients. Colors represent the main function of each gene, assigned on the bases of the information compiled in the GeneCards and KEGG databases. (B) Schematic representation of the mapping of CCND3 variants detected in SDRPL patients. (C) Details of each variant. BCR, B-cell receptor; cDNA, complementary DNA; CDS, coding sequence.
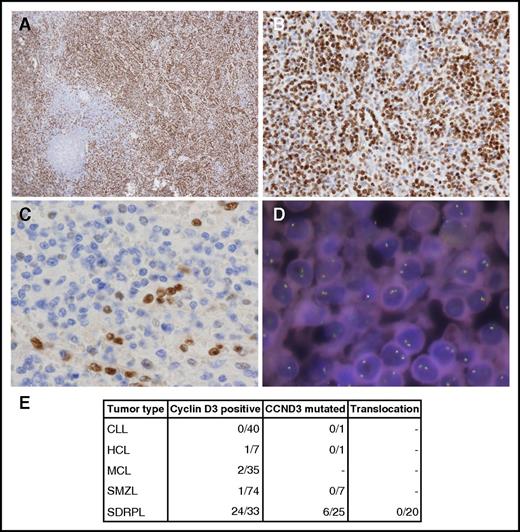
This evidence prompted us to explore the coding sequence of CCND3 in an extended series of splenic lymphomas that included SDRPL, SMZL, HCL, and chronic lymphocytic leukemia (CLL) cases. The last 2 exons of CCND3 of 34 patients (including 18 of the 23 patients whose exomes had previously been sequenced) were sequenced by polymerase chain reaction–based methods in a MiSeq instrument (supplemental Methods). The 4 variants found by WES were validated and 3 new variants in 2 patients were found (Figure 1B-C). Six of 25 SDRPL patients (24%) harbored mutations in CCND3. All were missense variants that fall into the proline, glutamic acid, serine, and threonine (PEST) domain (T283, P284, and I290), except 1 that was located at amino acid 202 (Figure 1B-C). Stabilization and increased expression of cyclin D3 has been described as a consequence of mutations in the PEST domain. Specifically, amino acids T283, P284, and I290 are part of a phosphorylation motif that regulates cyclin D3 phosphorylation and stability.17-19 Therefore, we decided to explore the expression of cyclin D3, which was investigated in 37 splenectomy specimens from SDRPL patients and was considered to be valuable (internal positive controls provided by the sinus-lining cells) in 33 specimens. Nonvaluable cases were fixed in Bouin. Twenty-four cases showed distinct cyclin D3 expression in >50% of the neoplastic cells, and 9 cases had a lower level of immunostaining, with cyclin D3 expression restricted to sinus-lining cells, megakaryocytes, and proliferating lymphoid cells. Cyclin D3 expression was also investigated in 74 SMZL, 35 mantle cell lymphoma (MCL), 7 HCL, and 40 CLL cases, with only 1 SMZL, 1 HCL, and 2 blastic MCL cases considered to be positive, whereas staining was apparent in most neoplastic cells (Figure 2).
Cyclin D3 in splenic B‐cell lymphomas. (A-B) 3,3′-diaminobenzidine (DAB) immunostaining of a cyclin D3-positive SDRPL case. Original magnification ×4 (A) and ×20 (B). (C) DAB immunostaining of a cyclin D3‐negative SMZL case. Original magnification ×40. (D) FISH staining with a cyclin D3 break-apart probe. DNA nuclei were stained with 4′,6-diamidino-2-phenylindole. Original magnification ×100. (E) Number of splenic B-cell lymphoma cases studied by cyclin D3 immunohistochemistry, CCND3 sequencing, and FISH analysis.
Cyclin D3 in splenic B‐cell lymphomas. (A-B) 3,3′-diaminobenzidine (DAB) immunostaining of a cyclin D3-positive SDRPL case. Original magnification ×4 (A) and ×20 (B). (C) DAB immunostaining of a cyclin D3‐negative SMZL case. Original magnification ×40. (D) FISH staining with a cyclin D3 break-apart probe. DNA nuclei were stained with 4′,6-diamidino-2-phenylindole. Original magnification ×100. (E) Number of splenic B-cell lymphoma cases studied by cyclin D3 immunohistochemistry, CCND3 sequencing, and FISH analysis.
Expression of cyclin D3 has previously been investigated in non-Hodgkin lymphomas (NHLs), in which cyclin D3 was present in a very low proportion of cases diagnosed as CLL (2 of 38), nodal MZL (1 of 5), MZL (0 of 24), SMZL (0 of 5), and MCL (1 of 5).20 More recently, cyclin D3 expression has been described in a blastoid variant of MCL with hypercalcemia.21
All the SDRPL cases of our series that harbored mutations in CCND3 showed high levels of the protein, but still there are 19 wild-type CCND3 cases showing high expression of cyclin D3. In order to explore a different mechanism that could explain this increased expression, we performed fluorescence in situ hybridization (FISH) analysis in 20 cyclin D3–positive cases using commercial probes against CCND3 (supplemental Methods). Cyclin D3 as a target gene of t(6;14)(p21.1;q32.3) of mature B-cell malignancies was initially described by Sonoki and coworkers in 2001 in a series of 6 NHL cases, 3 of which have clinical data consistent with the diagnosis of SDRPL and 1 of them (case 3) also had morphology consistent with the diagnosis of SRPL.22 Increased expression of cyclin D3 and t(6;14)(p21;q32) has been described in a small subgroup of cyclin D1–negative MCLs by Wlodarska and coworkers,23 but this finding has not been confirmed in other studies of cyclin D1–negative MCLs.24 The precise diagnosis of these cyclin D3–positive cases diagnosed as MCL should be revisited, considering the findings described here.
None of the 20 cases studied showed CCND3 translocation, suggesting that the expression of cyclin D3 could be regulated through alternative mechanisms, such as the activation of the Wnt/β-catenin pathway. Interestingly, a nonsense mutation in CDKN1B, the gene that encodes the cyclin-dependent kinase inhibitor p27 and that interacts with cyclin D3, was found in 1 patient. The same change was detected in 2 patients in a study of HCL in which CDKN1B was inactivated by mutations in 16% of the patients.25
In summary, our results indicate that cyclin D3 is expressed in the majority of SDRPL cases in contrast with other types of small B-cell lymphomas, thus facilitating the recognition of this disease. Increased expression is sometimes the result of somatic mutations in the PEST domain of the CCND3 gene. These findings allow the reinvestigation of the incidence and precise histological and clinical features of SDRPL cases. Our findings also suggest that there could be a potential therapeutic approach for these tumors involving the use of CDK4/CDK6 inhibitors.
The sequence data reported in this article have been deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/; accession number SRP093813).
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank the Biobank of Hospital Universitario Marqués de Valdecilla–IDIVAL (Santander, Spain) for providing biological samples.
This work was supported by grants from the Ministerio de Economía, Industria y Competitividad (RTICC RD06/0020/0107, RD12/0036/0060, PI 12/1682, PT13/0010/0007, PI16/01294, SAF2013-47416-R, CIBERONC-ISCIII, PIE15/0081) and the Asociación Española Contra el Cáncer, Spain. S.D. was supported by the Torres Quevedo subprogramme (Ministerio de Economía y Competitividad [MICINN]) under grant agreement PTQ-12-05391.
Contribution: S.C.-O. prepared the libraries for targeted sequencing, analyzed WES and targeted sequencing data, and wrote the manuscript; R. Mondéjar performed sequence analysis; C.A. and R. Marès prepared samples for sequencing; L.C. collected samples and clinical data; A.B. and S.G.d.V. performed FISH analysis; M.M., A.T.-G., A.V., L.B., F.I.C., K.S., A.X., T.P., G.K., M.G.-C., M.P., and A.W. provided clinical samples and performed diagnoses; Y.-C.M. performed sequencing analysis; S.D., S.B., and I.G. performed WES bioinformatics analysis; M.G. and J.B. performed sample and library preparations and WES; J.P.V. helped write the manuscript; M.A.P. designed the study, supervised the project, and helped write the manuscript; and N.M. designed the study, supervised the project, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nerea Martinez, Instituto de Investigación Marqués de Valdecilla–IDIVAL, Avda Cardenal Herrera Oria S/N, E-39011 Santander, Spain; e-mail: nmartinez@idival.org.
References
Author notes
M.A.P. and N.M. are senior authors.