Key Points
Mass spectrometry is a high-throughput, low-resource technique that can identify immunoglobulin variable region gene from tissue specimens.
IGVL gene usage is restricted and different between systemic and localized AL and only partially explains organ tropism in this disease.
Abstract
The goal of this study was to investigate the frequency of use of light-chain variable region (IGVL) genes among patients with systemic (ALS) and localized (ALL) amyloidosis and to assess for associations between IGVL gene usage and organ tropism. We evaluated clinic charts from 821 AL patients seen at the Mayo Clinic who had bone marrow, fat pad, and solid organ tissue samples typed by liquid chromatography tandem mass spectrometry (LC-MS). We identified 701 patients with ALS and 120 with ALL. Overall, we were able to identify an IGVL gene in 87 (72%) patients with ALL and 573 (82%) patients with ALS. When compared with ALL, LV6-57 was more common, whereas KV3-20 and heavy-chain codeposition were less common in ALS. In this large series of ALS, characteristics particular to specific genotypes became apparent. LV6-57 patients were more likely to have renal involvement and to harbor a translocation 11;14. LV3-01 patients were less likely to have advanced cardiac disease and renal involvement. LV2-14 patients were more likely to have peripheral nerve involvement, an intact circulating immunoglobulin, and lower circulating dFLC. LV1-44 patients were more likely to have cardiac involvement. KV1-33 patients had more liver involvement and higher circulating dFLC. Finally, KV1-05 was associated with inferior overall survival but not independently of cardiac stage. IGVL gene usage appears to provide clues about disease pathophysiology and tissue tropism. LC-MS is a high-throughput and low-resource technique that can be used to identify IGVL gene from clinical tissue specimens.
Introduction
Systemic immunoglobulin light-chain amyloidosis (ALS) is the most common form of systemic amyloidosis, and despite significant advances in the field, it remains a fatal disease with a worse prognosis compared with that of multiple myeloma.1,2 ALs is caused by the progressive deposition of structurally unstable, clonal immunoglobulin light chains that form systemic amyloid deposits. Outcomes of patients with AL amyloidosis are primarily dictated by the type and extent of organ involvement, especially cardiac involvement.3 Little is known about what regulates organ tropism in this disease.
Sometimes, amyloid deposition may be localized to the site of amyloid precursor protein production by a neoplastic plasma cell clone. These patients are labeled as having localized AL amyloidosis (ALL) and have a much better prognosis because vital organs like heart, liver, and kidney are not compromised. These patients do not require systemic treatment of their AL amyloidosis. For these reasons, accurate distinction between the two is very important.
In an effort to elucidate mechanisms of organ tropism in ALS, prior authors hypothesized that organ tropism may be a function of light-chain variable region (IGVL) gene and gene family of the clone.4-9 These contributions have been seminal because they helped establish important trends in IGVL gene rearrangements that confer a higher risk for specific organ involvement. These studies involved a small number of patients because they relied on the use of time-consuming, polymerase chain reaction–based sequencing of bone marrow plasma cells to identify IGVL genes and have not been translated to the clinical practice because of complexity of the analytical process.
We have established a proteomics-based clinical assay for identifying amyloid types using clinical tissue sample proteomic analysis by liquid chromatography/tandem mass spectrometry (LC-MS).10-12 This method has been in routine use in a CAP/CLIA-compliant clinical laboratory in our institution since 2008. This method was expanded to identify clonotypic peptides from AL deposits and identify the IGVL gene and gene family with 100% specificity.11 Here we apply this methodology in the largest cohort of patients with ALs and ALL in an effort to definitively characterize the relationship between the IGVL gene repertoire and organ tropism in this disease. We propose that the ability to identify the amyloidogenic clone directly from clinical tissue specimens using a proteomics-based methodology is a novel and potentially powerful clinical and research tool.
Methods
Patient population and clinical data
We included patients with ALL and ALS seen at the Mayo Clinic between August of 1991 and July of 2013. The Mayo Foundation Institutional Review Board approved the study and only patients who consented to having their medical records reviewed for research purposes were included. The diagnosis of AL amyloidosis was predicated on the presence of a biopsy specimen that stained positive by Congo red and exhibited green birefringence under polarized light and was documented to be AL amyloidosis by typing with LC-MS. Individual patients’ records were reviewed to abstract clinical organ involvement and systemic vs localized involvement. Patients without a circulating monoclonal protein (detected by immunofixation of serum and urine and serum-free light-chain assay), a bone marrow without clonal plasma cells (detected by immunohistochemistry and/or flow cytometry), and a fat aspirate negative for Congo red–positive deposits were classified as ALL. Further tests to rule out systemic organ involvement in suspected ALL were done as clinically indicated. Organ involvement for ALS was defined using established criteria.13 All patients with cardiac involvement had either a positive endomyocardial biopsy or a suggestive echocardiographic picture in addition to elevation of at least one cardiac biomarker (ie, B-type natriuretic peptide [BNP], N-terminal pro B-type natriuretic peptide [NTproBNP], or troponin). No patient without biopsy-confirmed cardiac involvement had normal cardiac biomarkers. Isolated cardiac or renal involvement was defined as isolated involvement of only the heart or kidneys, respectively, without other organs being clinically involved. Accurate abstraction of absence of organ involvement was not possible for some patients based on the limited information provided in their medical records, leaving 679 (97%) patients with cardiac involvement and 664 (95%) patients with renal involvement evaluable for these analyses. Extent of cardiac involvement was graded according to the Mayo 2004 staging system,3 which defines stage I as troponin T <0.035 ng/mL and NT-proBNP <332 ng/mL, stage III as both biomarkers equal to or above the respective thresholds, and stage II as either biomarker equal to or above the respective thresholds. Extent of renal involvement was graded according to the recently published renal staging system.14
Identification of immunoglobulin variable region gene usage and heavy-chain codeposition
The proteomics method for amyloid typing has been previously described both for solid tissue specimens and fat aspirates.10-12 Briefly, for patients with solid tissue specimens, laser microdissection (LMD) was performed on 3 to 4 Congo red–positive areas of formalin-fixed paraffin-embedded tissue blocks. The dissected tissue was digested into tryptic peptides and analyzed by LC-MS. For each fat aspirate specimen, 3 to 4 spicules were individually solubilized and digested in trypsin without undergoing LMD.12 Tandem MS spectra present in each sample’s raw file were identified with 3 different database search engines: Sequest,15 X!Tandem,16 and Mascot,17 all of which had been augmented with known protein sequences from human IGVL genes and families. The method for identifying IGVL gene was validated in an independent population (n = 30) by comparing it with mRNA sequencing performed on bone marrow samples,11 and it was found to be 100% specific.
All peptides identified from a patient’s sample were combined and filtered using the Scaffold software (Proteome Software, Portland, OR). Peptide identifications were accepted if they could be established with a >95% probability, at a 90% confidence interval. A spectral counting approach was used, and a count ≥5 was considered to be clinically significant. For every case, we created a proteomics profile that lists all of the confident protein identifications present in each sample along with their respective spectral counts. The IGVL gene was identified based on abundance of spectral counts.
We also considered the presence or absence of heavy chains in tissue specimens. Because we do not have a robust method to identify contamination of samples by serum immunoglobulins, we excluded all IgG-class immunoglobulins for all heavy-chain analyses because these are found in high concentrations in human serum. We included all detectable IgM, IgA, and IgD types. Furthermore, because our sequence libraries are not enriched to allow identification of IGH variable genes, we only examined IGH as a dichotomous variable based on the presence or absence of IGH-constant regions in amyloid specimens.
Statistical analysis
The Fisher exact test and the Kruskal-Wallis test were used to ascertain differences between categorical and continuous variables, respectively. For gene groups that contained at least 30 patients, differences between those with and without the gene were sought for multiple characteristics including: presence or absence of organ involvement, plasma cell clone characteristics (dFLC, bone marrow plasma cells, fluorescence in situ hybridization [FISH]/cytogenetics), and differences in markers of organ damage (proteinuria, cardiac stage, renal stage). P values <.01 were considered significant to correct for multiple comparisons. P values between .01 and .05 were noted to be possible trends and worthy of future exploration with larger sample sizes of patients. Overall survival (OS) from diagnosis was calculated using the Kaplan-Meier method. All statistics were done using JMP software (SAS, Cary, NC).
Results
IGVL gene distribution and heavy-chain codeposition in ALL and ALS
The patients’ baseline characteristics for ALS and ALL are shown in Table 1. We identified 120 patients with ALL and 701 patients with ALS. Patients with ALL were younger and less likely to be male. The number of κ ALL and ALS were 54 (45%) and 187 (27%), respectively. Overall, we were able to identify an IGVL gene in 86 (72%) patients with ALL and in 573 (82%) patients with ALS (P = .01). There was no difference in identifying IGVL gene between κ- and λ-restricted cases. We could not identify an IGVL gene or family in 16 (13%) ALL cases and 79 (11%) ALs cases (Figure 1).
Demographics and clinical characteristics of patients
| Characteristic (N = 821) . | ALS (N = 701) . | ALL (N = 120) . |
|---|---|---|
| Male | 445 (64%) | 46 (38%) |
| Age (y), median (range) | 63 (36-89) | 60 (32-86) |
| Seen at Mayo within 90 d of diagnosis | 530 (76%) | 61 (50%) |
| Circulating monoclonal protein in the serum or urine* | ||
| κ | 187 (27%) | |
| IgG | 270 (39%) | |
| IgA | 40 (6%) | |
| IgM | 42 (6%) | |
| IgD | 5 (0.7%) | |
| Light chain only | 284 (41%) | |
| Biclonal | 5 (0.7%) | |
| No circulating monoclonal protein | 18 (3%) | |
| No data on monoclonal protein | 37 (5%) | |
| Organ involvement | ||
| Cardiac | 457 (65%) | 0 |
| Mayo 2004 cardiac stage† | NA | |
| I | 76 (19%) | |
| II | 150 (38%) | |
| III | 169 (43%) | |
| Renal | 357 (51%) | 0 |
| Renal stage‡ | NA | |
| I | 64 (42%) | |
| II | 70 (46%) | |
| III | 17 (12%) | |
| Peripheral and autonomic nervous system | 112 (16%) | 0 |
| Hepatic | 56 (8%) | 0 |
| Gastrointestinal system | 174 (25%) | 18 (16%) |
| Skin | 23 (3%) | 36 (30%) |
| Other soft tissue | 100 (14%) | 37 (31%) |
| Lung parenchymal | 43 (6%) | 31 (26%) |
| Lymph nodes | 13 (2%) | 1 (0.8%) |
| Bladder or ureter | 5 (1%) | 18 (15%) |
| Tracheobronchial | 0 | 5 (4%) |
| Characteristic (N = 821) . | ALS (N = 701) . | ALL (N = 120) . |
|---|---|---|
| Male | 445 (64%) | 46 (38%) |
| Age (y), median (range) | 63 (36-89) | 60 (32-86) |
| Seen at Mayo within 90 d of diagnosis | 530 (76%) | 61 (50%) |
| Circulating monoclonal protein in the serum or urine* | ||
| κ | 187 (27%) | |
| IgG | 270 (39%) | |
| IgA | 40 (6%) | |
| IgM | 42 (6%) | |
| IgD | 5 (0.7%) | |
| Light chain only | 284 (41%) | |
| Biclonal | 5 (0.7%) | |
| No circulating monoclonal protein | 18 (3%) | |
| No data on monoclonal protein | 37 (5%) | |
| Organ involvement | ||
| Cardiac | 457 (65%) | 0 |
| Mayo 2004 cardiac stage† | NA | |
| I | 76 (19%) | |
| II | 150 (38%) | |
| III | 169 (43%) | |
| Renal | 357 (51%) | 0 |
| Renal stage‡ | NA | |
| I | 64 (42%) | |
| II | 70 (46%) | |
| III | 17 (12%) | |
| Peripheral and autonomic nervous system | 112 (16%) | 0 |
| Hepatic | 56 (8%) | 0 |
| Gastrointestinal system | 174 (25%) | 18 (16%) |
| Skin | 23 (3%) | 36 (30%) |
| Other soft tissue | 100 (14%) | 37 (31%) |
| Lung parenchymal | 43 (6%) | 31 (26%) |
| Lymph nodes | 13 (2%) | 1 (0.8%) |
| Bladder or ureter | 5 (1%) | 18 (15%) |
| Tracheobronchial | 0 | 5 (4%) |
Identified by immunofixation of serum or urine or by the serum-free light-chain assay.
Of 701 patients, 395 (56%) had sufficient data available to assign cardiac stage.
Of 350 patients with renal involvement, 151(43%) had enough data available to assign renal stage.
Immunoglobulin variable region gene usage in systemic and localized amyloidosis.KV1-other: includes rare KV1-genes (KV1-02, 06, 08, 09, 12, 13, 17, and 27) and cases where only KV1 family could be determined. Other κ/λ: includes rare κ genes (n = 8, KV2-28, 30, 38, KV3-01, 07, 11, KV6-21, rare KV1 excluded because included in KV1-other); rare λ genes (n = 29, LV1-01, 36, 52, 57, LV2-08, 11, 18, LV3-09, 10, 20, 25, LV4-60, 69, and LV10-54), cases where only family could be determined (n = 49); IGKC and IGLC: cases that no gene or family could be determined (n = 86). HC, non-IgG heavy chain. *P < .05, **P < .01, ***P < .001.
Immunoglobulin variable region gene usage in systemic and localized amyloidosis.KV1-other: includes rare KV1-genes (KV1-02, 06, 08, 09, 12, 13, 17, and 27) and cases where only KV1 family could be determined. Other κ/λ: includes rare κ genes (n = 8, KV2-28, 30, 38, KV3-01, 07, 11, KV6-21, rare KV1 excluded because included in KV1-other); rare λ genes (n = 29, LV1-01, 36, 52, 57, LV2-08, 11, 18, LV3-09, 10, 20, 25, LV4-60, 69, and LV10-54), cases where only family could be determined (n = 49); IGKC and IGLC: cases that no gene or family could be determined (n = 86). HC, non-IgG heavy chain. *P < .05, **P < .01, ***P < .001.
KV1-33 was the most common κ IGVL, and the KV1 family overall was the most common κ family identified in both ALS (16% of cases) and ALL (12% of cases) (Figure 1). KV3-20 was underrepresented in ALS compared with ALL (1% vs 9%, respectively; P < .001), and LV6-57 was overrepresented (15% vs 5%, respectively; P < .001), in addition to being the most common IGVL gene in ALS. Overall, the LV6-57, LV3-01, and LV2-14 genes and the KV1 family constituted 49% of clones identified in ALS.
Non-IgG heavy chains (IGH) were present in the amyloid deposits of 14 (12%) patients with ALL (5 IgA, 3 IgD, and 6 IgM) and 11 (2%) of patients with ALs (1 IgA, 10 IgM) (P < .001) (Figure 1). Of the 11 systemic patients, 7 had a circulating heavy chain with the same restriction as the one deposited (6 with IgM and 1 with IgA). To exclude biopsy site as a source of bias, we examined heavy-chain codeposition across various tissue types. The proportion of samples with heavy-chain codeposition in ALS was similar among bone marrow, fat aspirate, and nonmarrow/fat samples.
IGVL usage and organ tropism
Although this is the largest series of patients of AL IGVL, given the diversity of the repertoire of gene use in patients with AL, the actual numbers of patients using any given IGVL is small. The groups with at least 30 patients are shown in Table 2. As mentioned in the methods, P values <.01 were considered significant, but P values <.05 were considered highly suggestive of potential trends. Of 701 ALS patients, 679 (97%) had enough information to ascertain presence or absence of cardiac involvement and 664 (95%) to ascertain presence or absence of renal involvement.
Notable patient characteristics by IGVL gene usage for most abundant clones*
| Gene . | Organ involvement . | dFLC levels/amyloidogenicity . | Organ toxicity . | FISH abnormalities . |
|---|---|---|---|---|
| LV6-57 (n = 102) | ↑ Renal involvement: 79% vs 48%, P = .0004 | No differences | 24-h urine protein >5 g 56% vs 34%, P = .02† | ↑t(11;14) 55% vs 40%, P < .05↓Trisomies 9.5% vs 22%, P < .05 |
| LV3-01 (n = 66) | ↓ Renal involvement: 41% vs 54%, P < .05 | No differences | ↓ Mayo stage III, 26% vs 43%, P = .02† | No differences |
| LV2-14 (n = 64) | ↑ In gastrointestinal ALL | ↓ dFLC 9.47 vs 24.5 mg/dL, P = .025 | No differences | No differences |
| ↑ Peripheral nerve involvement: 25% vs 14%, P < .05 | ↑ Intact circulating Ig 80% vs 57%, P = .002 | |||
| LV1-44 (n = 45) | ↑ Cardiac involvement: 80% vs 65%, P = .03 | No differences | No differences | ↓Trisomies 4% vs 22%, P < .05 |
| KV1-33 (n = 40) | ↑ Liver involvement 27% vs 7%, P = .0002 | ↑ dFLC 64.8 mg/dL vs 22.2 mg/dL, P < .002 | No differences | No differences |
| ↓ Peripheral nerve involvement 2.7% vs 15.6%, P = .017 | ||||
| LV3-21 (n = 38) | ↓ Renal involvement 29% vs 54%, P = .0025 | No differences | ↑ Monosomy 13q/13q 61% vs 33%, P = .017 |
| Gene . | Organ involvement . | dFLC levels/amyloidogenicity . | Organ toxicity . | FISH abnormalities . |
|---|---|---|---|---|
| LV6-57 (n = 102) | ↑ Renal involvement: 79% vs 48%, P = .0004 | No differences | 24-h urine protein >5 g 56% vs 34%, P = .02† | ↑t(11;14) 55% vs 40%, P < .05↓Trisomies 9.5% vs 22%, P < .05 |
| LV3-01 (n = 66) | ↓ Renal involvement: 41% vs 54%, P < .05 | No differences | ↓ Mayo stage III, 26% vs 43%, P = .02† | No differences |
| LV2-14 (n = 64) | ↑ In gastrointestinal ALL | ↓ dFLC 9.47 vs 24.5 mg/dL, P = .025 | No differences | No differences |
| ↑ Peripheral nerve involvement: 25% vs 14%, P < .05 | ↑ Intact circulating Ig 80% vs 57%, P = .002 | |||
| LV1-44 (n = 45) | ↑ Cardiac involvement: 80% vs 65%, P = .03 | No differences | No differences | ↓Trisomies 4% vs 22%, P < .05 |
| KV1-33 (n = 40) | ↑ Liver involvement 27% vs 7%, P = .0002 | ↑ dFLC 64.8 mg/dL vs 22.2 mg/dL, P < .002 | No differences | No differences |
| ↓ Peripheral nerve involvement 2.7% vs 15.6%, P = .017 | ||||
| LV3-21 (n = 38) | ↓ Renal involvement 29% vs 54%, P = .0025 | No differences | ↑ Monosomy 13q/13q 61% vs 33%, P = .017 |
BMPC, bone marrow plasma cells.
Characteristics tested for significance included: dFLC, circulating intact immunoglobulin, bone marrow plasma cells, presence of t(11;14), deletion 13q or trisomies by FISH, renal involvement, cardiac involvement, nerve involvement, liver involvement, Mayo 2004 cardiac stage, urine total protein, and estimated glomerular filtration rate.
Comparison group for each row is patients not harboring this IGVL gene.
Independent of baseline dFLC levels.
Not surprisingly, renal involvement was more common in patients with LV6-57 compared with those without, and was less common in patients with LV3-21 compared with those without (Table 2). When considering less common genes, KV1-05 patients had a trend toward less renal involvement compared with those without: 18% vs 53%, P = .02. IGH deposition was not different between patients with and without renal involvement. When examining the 78 patients with and without isolated renal involvement, no significant difference in IGVL gene usage or heavy-chain codeposition was found.
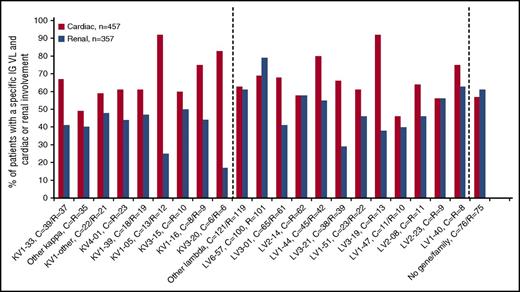
Cardiac involvement was very prevalent and was present in 66% of patients. In spite of this, the likelihood of cardiac involvement was particularly high among patients with certain genes: 92% for patients with LV3-19 and 92% for patients with KV1-05 (Figure 2). However, these genes were identified in only 2% of ALs cases. The rate of cardiac involvement for patients with these genes and with LV1-44 (Table 2) compare to those without and were as follows: LV3-19 (92% vs 65%, P = .02), KV1-05 (92% vs 65%, P = .03), and LV1-44 (80% vs 65%, P = .03; Table 2). IGH deposition was not different between patients with and without cardiac involvement. When comparing the 84 patients with isolated cardiac involvement (ie, no other organ involvement) to those without (ie, with cardiac but also with other organ involvement), no significant difference in IGVL gene usage or heavy-chain codeposition was found.
Cardiac and renal involvement according to immunoglobulin variable region gene usage. Other κ/λ: includes rare κ genes (KV2-28, 30, 38, KV3-01, 07,11, KV6-21, rare KV1 excluded); rare λ genes (LV1-01, 36, 52, 57, LV2-08, 11, 18, LV3-09, 10, 20, 25, LV4-60, 69, and LV10-54); cases where only family could be determined (n = 39); and cases that no gene or family could be determined (n = 77); KV1-other: includes rare KV1-genes (KV1-02, 06, 08, 09,12, 13, 17 and 27). P = nonsignificant across groups within patients with cardiac involvement. P < .0001 across gene groups within patients with renal involvement. C, cardiac; R, renal.
Cardiac and renal involvement according to immunoglobulin variable region gene usage. Other κ/λ: includes rare κ genes (KV2-28, 30, 38, KV3-01, 07,11, KV6-21, rare KV1 excluded); rare λ genes (LV1-01, 36, 52, 57, LV2-08, 11, 18, LV3-09, 10, 20, 25, LV4-60, 69, and LV10-54); cases where only family could be determined (n = 39); and cases that no gene or family could be determined (n = 77); KV1-other: includes rare KV1-genes (KV1-02, 06, 08, 09,12, 13, 17 and 27). P = nonsignificant across groups within patients with cardiac involvement. P < .0001 across gene groups within patients with renal involvement. C, cardiac; R, renal.
When considering organs other than heart and kidneys, there was a trend for patients with LV2-14 to have a higher rate of peripheral neuropathy (Table 2). KV1-33 was more likely to be associated with liver involvement and less likely to be associated with peripheral nerve involvement (Table 2).
Among the 120 patients with ALL, LV2-14 was overrepresented in those with gastrointestinal involvement (Table 3), but otherwise no other statistically significant relationships were identified including non-IgG heavy-chain codeposition.
Light-chain variable region gene distribution in localized amyloidosis according to organ involvement
| Ig V gene . | Skin, N = 36 . | Lung parenchyma, N = 31 . | Urothelial, N = 18 . | Gastrointestinal, N = 17 . |
|---|---|---|---|---|
| KV1-33 | 2 (6%) | 1 (3%) | 0 | 2 (11%) |
| Other κ | 5 (14%) | 5 (16%) | 0 | 2 (11%) |
| KV1-other | 1 (3%) | 0 | 0 | 0 |
| KV4-01 | 1 (3%) | 1 (3%) | 2 (11%) | 1 (5%) |
| KV1-39 | 3 (8%) | 0 | 0 | 0 |
| KV1-05 | 3 (8%) | 1 (3%) | 0 | 0 |
| KV3-15 | 1 (3%) | 1 (3%) | 2 (11%) | 1 (5%) |
| KV1-16 | 1 (3%) | 0 | 0 | 0 |
| KV3-20 | 2 (6%) | 5 (16%) | 1 (6%) | 2 (11%) |
| Other λ | 6 (17%) | 8 (26%) | 5 (28%) | 1 (5%) |
| LV6-57 | 0 | 3 (10%) | 2 (11%) | 0 |
| LV3-01 | 2 (6%) | 1 (3%) | 0 | 1 (5%) |
| LV2-14* | 2 (6%) | 0 | 2 (11%) | 5 (26%) |
| LV1-44 | 1 (3%) | 1 (3%) | 1 (6%) | 1 (1%) |
| LV3-21 | 2 (6%) | 0 | 0 | 1 (5%) |
| LV1-51 | 1 (3%) | 2 (7%) | 0 | 1 (5%) |
| LV3-19 | 0 | 1 (3%) | 0 | 0 |
| LV1-47 | 1 (3%) | 0 | 1 (6%) | 0 |
| LV2-08 | 0 | 0 | 1 (6%) | 0 |
| LV2-23 | 1 (3%) | 0 | 0 | 0 |
| LV1-40 | 1 (3%) | 1 (3%) | 1 (6%) | 0 |
| No gene/family | 1 (3%) | 3 (10%) | 4 (22%) | 1 (5%) |
| Codeposited heavy chain | 2 (6%) | 1 (3%) | 0 | 2 (11%) |
| Ig V gene . | Skin, N = 36 . | Lung parenchyma, N = 31 . | Urothelial, N = 18 . | Gastrointestinal, N = 17 . |
|---|---|---|---|---|
| KV1-33 | 2 (6%) | 1 (3%) | 0 | 2 (11%) |
| Other κ | 5 (14%) | 5 (16%) | 0 | 2 (11%) |
| KV1-other | 1 (3%) | 0 | 0 | 0 |
| KV4-01 | 1 (3%) | 1 (3%) | 2 (11%) | 1 (5%) |
| KV1-39 | 3 (8%) | 0 | 0 | 0 |
| KV1-05 | 3 (8%) | 1 (3%) | 0 | 0 |
| KV3-15 | 1 (3%) | 1 (3%) | 2 (11%) | 1 (5%) |
| KV1-16 | 1 (3%) | 0 | 0 | 0 |
| KV3-20 | 2 (6%) | 5 (16%) | 1 (6%) | 2 (11%) |
| Other λ | 6 (17%) | 8 (26%) | 5 (28%) | 1 (5%) |
| LV6-57 | 0 | 3 (10%) | 2 (11%) | 0 |
| LV3-01 | 2 (6%) | 1 (3%) | 0 | 1 (5%) |
| LV2-14* | 2 (6%) | 0 | 2 (11%) | 5 (26%) |
| LV1-44 | 1 (3%) | 1 (3%) | 1 (6%) | 1 (1%) |
| LV3-21 | 2 (6%) | 0 | 0 | 1 (5%) |
| LV1-51 | 1 (3%) | 2 (7%) | 0 | 1 (5%) |
| LV3-19 | 0 | 1 (3%) | 0 | 0 |
| LV1-47 | 1 (3%) | 0 | 1 (6%) | 0 |
| LV2-08 | 0 | 0 | 1 (6%) | 0 |
| LV2-23 | 1 (3%) | 0 | 0 | 0 |
| LV1-40 | 1 (3%) | 1 (3%) | 1 (6%) | 0 |
| No gene/family | 1 (3%) | 3 (10%) | 4 (22%) | 1 (5%) |
| Codeposited heavy chain | 2 (6%) | 1 (3%) | 0 | 2 (11%) |
Other κ/λ: includes rare κ genes (KV2-28, 30, 38, KV3-01, 07, 11, KV6-21, rare KV1 excluded); rare λ genes (LV1-01, 36, 52, 57, LV2-08, 11, 18, LV3-09, 10, 20, 25, LV4-60, 69, and LV10-54); cases where only family could be determined; and cases that no gene or family could be determined. KV1-other: includes rare KV1-genes (KV1-02, 06, 08, 09, 12, 13, 17, and 27).
P < .01.
IGVL association with amyloidogenicity and plasma cell clone characteristics
Among other variables, we examined whether there were any differences in baseline dFLC levels according to IGVL gene, hypothesizing that more amyloidogenic clones could cause clinical organ involvement at lower dFLC levels compared with less amyloidogenic ones. Of 701 ALS patients, 463 (66%) had dFLC levels available at diagnosis. Finally, because bone marrow FISH has been associated with organ involvement and prognosis in ALS,18-20 we examined whether there was any differential IGVL gene usage according to bone marrow FISH.
Patients with KV1-33 were more likely to have higher and patients with LV2-14 lower baseline dFLC levels, respectively, compared with patients without these IGVL genes (Table 2). Patients with a LV2-14–producing clone were also more likely to have an intact circulating immunoglobulin (Table 2). There were no differences in IGVL gene usage between patients with very low dFLC levels (<5 mg/dL) and patients with dFLC (≥5mg/dL).
We noted that patients with LV6-57 were more likely to have more advanced renal disease (24-hour urine protein >5 gr) independently of baseline dFLC levels (odds ratio [OR] 2.5, 95% confidence interval [CI] 1.2-5.4; P = .02). Patients with LV3-01 had a trend toward lower (OR 2.6, 95% CI 1.1-6.6; P = .03) and patients with KV1-05 had a trend toward higher (OR 4.4, 95% CI 1.03-30; P = .045) rates of stage III cardiac disease, respectively, an effect independent of baseline dFLC levels.
There was a trend for patients with LV6 to be more likely to harbor the t(11;14) translocation and less likely to harbor trisomies. When considering patients with LV1-44, trisomies appeared to be less common (Table 2). Finally, LV3-21 patients had a trend of higher bone marrow plasmacytosis and higher frequency of monosomy 13 (Table 2).
IGVL and overall survival
We performed limited exploratory analyses evaluating IGVL usage and OS mostly because we felt we there were a lot of missing data to draw safe conclusions in regards to OS, which is influenced by several factors we were not able to accurately capture in this study (hematologic response, organ response, host factors, delayed diagnosis). Of 701 patients with ALS, first-line treatment data were available for 385 (55%) patients, hematologic response to first-line treatment was available for 361 (51%) patients, and only 103 (23%) patients with cardiac involvement and 143 (40%) patients with renal involvement had at least 12 months of follow-up for organ response.
Only KV1-05 appeared to be associated with a decreased OS compared with patients harboring other IGVL genes: median survival 12 months vs 59 months, P = .003 (supplemental Figure 1, available on the Blood Web site). This effect, however, was not independent of cardiac stage on a multivariate model including KV1-05, IGVL type and cardiac stage.
Discussion
This is the largest study to date reporting on IGVL gene and family usage in patients with systemic and localized AL amyloidosis. For the first time, we demonstrate that IGVL gene usage is different between ALS and ALL and that non-IgG heavy-chain deposition is more common in ALL compared with ALS. We identify new potential predilections for IGVL gene usage in ALS patients with cardiac, renal, and hepatic involvement, and ALL patients with gastrointestinal involvement. Finally, we identify new clones that are more likely to be associated with different levels of organ damage in ALS.
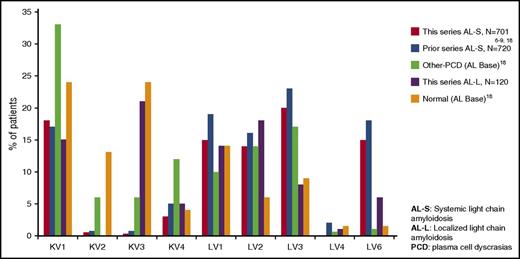
Figure 3 provides a comparison between our results and those that have been published in the literature,6-9 and those that can be found in AL Base.21 Of the 29 to 33 functional IGV λ genes and the 31 to 36 functional IGV κ genes in the human genomic repertoire, consistent with previous publications, LV6-57 is the most common IGVL gene identified among patients with ALs.8,22 Approximately half of the light-chain variable gene repertoire in ALS is comprised of LV6-57, LV3-01, and LV2-14 genes and the KV1 family (particularly KV1-33). Not only are these genes the most commonly observed in ALS, but they and their gene families are also conspicuously more common in ALS than in the normal B-repertoire.21,23,24 In contrast, the distribution of IGVL genes and families for patients with ALL was very similar to that seen in the normal B-cell repertoire, with LV6 found less commonly and the KV3 family (and the KV3-20 gene in particular) more commonly.8 The potential mechanisms underlying the differences between IGVL gene usage in ALS, ALL, nonamyloidogenic plasma cell dyscrasias, and normal controls are not clear. In the case of ALL, the bias of IGVL gene usage might be explained by different antigenic stimuli. ALL is often associated with autoimmune diseases25 and lymphoproliferative disorders that may be autoimmune driven (ie, MALT lymphomas).26
Variable region family usage in light-chain amyloidosis, other plasma cell dyscrasias, and normal subjects. Data for the prior ALs series are pooled from references 6,,-9,18 and data for both the other plasma cell dyscrasias and normal B cells are derived from AL Base.21
When considering patients with cardiac involvement, some of our results are consistent with those of Perfetti and colleagues.9 We also found a greater than twofold higher rate of cardiac involvement among patients with LV1-44. In their study of 99 patients with ALS, Perfetti and colleagues observed trends for a lower risk for patients with LV3-01 and LV6-57 to have “dominant” cardiac involvement.9 In our study, rates of cardiac involvement for these two genotypes were no different from those found in other genotypes, but patients with LV3-01 were more likely to have less severe cardiac involvement by Mayo stage. Our study also revealed provocative trends among cardiac patients that will need to be reproduced in larger cohorts. For example, >90% of patients with KV1-05 and LV3-19 had cardiac involvement; the significance of this finding is tempered by the fact that there were only 16 and 13 patients with these variable genes in our systemic cohort. KV1-05 was also the only IGVL associated with worse overall survival, although this effect appeared to be dependent on cardiac stage.
In patients with renal involvement, LV6-57 was more commonly encountered, which is similar to prior reports.6,7 In addition, our observations that renal involvement was less common in patients with LV3-01 and LV3-21 are novel. Furthermore, LV6-57 was associated with higher levels of proteinuria independent of baseline dFLC levels, suggesting that LV6-57 might be more nephrotoxic. LV6-57 was more commonly associated with translocation 11;14, which is the most common FISH abnormality found in AL amyloidosis but also in other monoclonal gammopathies of renal significance27 and multiple myeloma patients presenting with renal damage.28
Other interesting novel observations included increased and decreased rates of peripheral neuropathy in patients with LV2-14 and KV1-33, respectively. Finally, patients with hepatic involvement have previously been shown to be more commonly κ restricted,6,29 although the dominance of KV1-33 is a novel finding in our study.
In general, the KV1 family appeared to be less amyloidogenic in vivo, because it required higher levels of circulating free light chains (ie, dFLC) to lead to clinically significant organ involvement. This has been supported by biophysical experiments.30-32 In contrast, some of the increased amyloidogenicity seen with LV2-14 could be attributed to the higher burden of nonconservative mutations—which have been associated with increased amyloidogenicity—within the λ family.33,34 Our results regarding extent of organ involvement and IGVL amyloidogenicity are novel and potentially hypothesis-generating but at the same time exploratory and require further validation.
In our study, heavy chains were more commonly codeposited in ALL. Furthermore, our study probably underestimated the codeposition of IGH in ALL, because we had to exclude all IgG-class IGH for the lack of a robust mechanism to exclude sample contamination by serum IgG. Nonetheless, IGH codeposition has been noted in both ALL and ALS in the past.26,35-38 It needs to be noted, however, that we could not identify the variable heavy-chain region in codeposited IGH, and therefore we could not differentiate between monoclonal or polyclonal IGH deposits. The fact that in most ALS patients the heavy chain isolated in the amyloid deposits was also detected in the serum suggests that a systemic deposition mechanism exists at least for the IgM and IgA immunoglobulin subclasses in ALS.
Our results suggest that the IGVL gene and family restriction is only part of the puzzle that constitutes clinical organ tropism and amyloid toxicity. Other, as yet unidentified, factors, either intrinsic to the structure of the light-chain protein or related to the microenvironment of the amyloid plaque,35 as well as the composition of the codeposited proteins within the amyloid plaque itself, have also been postulated to play a role and should be the focus of future research.
Limitations of our study include the small number of patients harboring specific IGVL clones, which limits the power of some of our observations. Furthermore, there is still room for improvement in terms of the sensitivity of this assay, which will improve as our IGVL sequence template libraries and peptide identification algorithms improve. The number of cases included in this study is proven to be powerful enough to uncover significant trends, but further study will be required. Extent of organ involvement and amyloid toxicity is a function of other factors in addition to baseline dFLC, which could not always be captured with accuracy in this retrospective study (delay in diagnosis, host factors). Finally, we did not perform comprehensive OS or any organ response analyses primarily because of missing data. Furthermore, organ response and survival are influenced by several variables that we were not able to accurately capture in this retrospective study (eg, depth and especially duration of hematologic response, host factors, disease stage at presentation). Some of these exploratory analyses have been published in abstract form and no association was found.39
Strengths of the study include the size of the cohort, the inclusion of a large number of ALL patients, as well as the methodology itself, which is a fast and inexpensive method to evaluate IGVL genes from data already generated as part of the clinical typing assay.
In conclusion, we have clarified IGVL gene and family usage in a large cohort of patients with systemic and localized AL amyloidosis, using a novel methodology that relies on MS of clinical tissue specimens. We show for the first time that IGVL gene and family repertoire is different between ALS and ALL. Our data show that IGVL gene family usage does not entirely explain clinical organ tropism, but they can provide a platform upon which future research exploring how other aspects of the amyloid proteome relate to organ tropism and clinical outcome. Moreover, our observations about propensity of certain genes being more amyloidogenic and associated with particular presentations could have direct clinical implications, especially as the methodology for top down sequencing of circulating monoclonal proteins moves into the mainstream. For example, individuals with an apparent monoclonal gammopathy of undetermined significance with KV3 gene usage are unlikely to have or develop ALS; in contrast, patients with LV6-57 monoclonal gammopathy should be comprehensively evaluated for ALS and followed closely, because LV6-57 is more common in ALS than in the normal B-cell repertoire. In this fashion, the results presented in this study could be applied in ALS prevention, a major bottleneck in improving patient outcomes.40
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.V.K., S.D., P.J.K., and A.D. conceived and designed the study; T.V.K., S.D., and A.D. analyzed and interpreted data; and all authors provided study materials of patients, collected and assembled data, and wrote and gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests
Correspondence: Angela Dispenzieri, Division of Hematology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: dispenzieri.angela@mayo.edu.