Key Points
Knockdown of CDKI p19INK4d impairs human terminal erythroid differentiation by decreasing GATA1 protein levels.
GATA1 protein level is regulated by p19INK4d via the PEBP1-p-ERK-HSP70-GATA1 pathway.
Abstract
Terminal erythroid differentiation is tightly coordinated with cell-cycle exit, which is regulated by cyclins, cyclin-dependent kinases, and cyclin-dependent kinase inhibitors (CDKI), yet their roles in erythropoiesis remain to be fully defined. We show here that p19INK4d, a member of CDKI family, is abundantly expressed in erythroblasts and that p19INK4d knockdown delayed erythroid differentiation, inhibited cell growth, and led to increased apoptosis and generation of abnormally nucleated late-stage erythroblasts. Unexpectedly, p19INK4d knockdown did not affect cell cycle. Rather, it led to decreased expression of GATA1 protein. Importantly, the differentiation and nuclear defects were rescued by ectopic expression of GATA1. Because the GATA1 protein is protected by nuclear heat shock protein family (HSP) member HSP70, we examined the effects of p19INK4d knockdown on HSP70 and found that p19INK4d knockdown led to decreased expression of HSP70 and its nuclear localization. The reduced levels of HSP70 are the result of reduced extracellular signal-regulated kinase (ERK) activation. Further biochemical analysis revealed that p19INK4d directly binds to Raf kinase inhibitor PEBP1 and that p19INK4d knockdown increased the expression of PEBP1, which in turn led to reduced ERK activation. Thus we have identified an unexpected role for p19INK4d via a novel PEBP1-p-ERK-HSP70-GATA1 pathway. These findings are likely to have implications for improved understanding of disordered erythropoiesis.
Introduction
Erythropoiesis is a process during which hematopoietic stem cells are first committed to erythroid progenitors, which subsequently undergo terminal erythroid differentiation to produce mature red blood cells. In the process of terminal erythroid differentiation, proerythroblasts undergo 4 to 5 mitoses to sequentially generate basophilic erythroblasts, polychromatic erythroblasts, and orthochromatic erythroblasts that expel their nuclei to produce enucleated reticulocytes.1
Erythropoiesis is a tightly regulated process. The most well-studied regulatory mechanisms include EPO/EPOR-mediated signal transduction and transcription factors that regulate erythroid gene expression.2 The critical role of the transcription factor GATA1 has been well-established.2-4 Either deficiency or overexpression of GATA1 impairs erythropoiesis, leading to severe anemia.5-7 GATA1 expression can be regulated at both transcriptional and posttranscriptional levels.8-11 At the posttranscriptional level, GATA1 is mainly regulated by the heat shock protein (HSP) family members HSP27 and HSP70 and ribosomal proteins.9-11 HSP27 promotes GATA1 ubiquitination and proteasomal degradation,9 HSP70 prevents the caspase-3–mediated cleavage of GATA1 in the nucleus,10 and ribosomal proteins regulate the normal translation of GATA1.11 It has been shown that defective HSP70 nuclear localization is associated with GATA1 cleavage and ineffective erythropoiesis in myelodysplastic syndromes (MDSs) and thalassemias.5,6 Ribosomal protein haploinsufficiency reduces GATA1 translation and has been implicated in the pathogenesis of some Diamond-Blackfan anemia patients.11
Erythropoiesis, particularly terminal erythroid differentiation, is tightly associated with cell-cycle exit.12,13 Cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (CDKIs) regulate cell cycle. In general, cyclins and CDKs form cyclin–CDK complexes to drive cell-cycle progression,14 whereas CDKI promotes cell-cycle arrest by inhibiting the activity of CDK.15 Indeed, the roles of some cell-cycle regulators in erythropoiesis have been previously reported. For example, it has been shown that cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number,16 whereas cyclin A2 is a regulator of cytokinesis during terminal erythroid differentiation.17 CDKI consists of 2 families: the INK4 family that includes p15INK4b, p16INK4a, p18INK4c, and p19INK4d, and the CIP/KIP family that includes p21CIP1/WAF1, p27KIP1, and p57KIP2.18 Although previous studies have suggested roles for some of these CDKI family members in murine erythropoiesis,13,19-22 the expression and function of these family members in human terminal erythroid differentiation remains largely unexplored.
In the present study, we first showed that of the 7 CDKI family members, only p18INK4c, p19INK4d, and p27KIP1 are expressed in human erythroid cells and their expression is significantly upregulated in late stages of terminal erythroid differentiation. Because the roles of p18INK4c and p27KIP1 in murine erythropoiesis have been previously described,13,19,22 we focused our efforts on exploring the function of p19INK4d on human erythropoiesis. We unexpectedly found that p19INK4d plays an important role in human terminal erythropoiesis by maintaining GATA1 protein levels through a novel PEBP1-p-ERK-HSP70 pathway. These findings likely have implications for improved understanding of disordered erythropoiesis in human disorders such as thalassemias, MDSs, and congenital dyserythropoietic anemias.
Materials and methods
Reagents and antibodies
Extracellular signal-regulated kinase (ERK) inhibitor (FR180204) was obtained from Selleck (Houston, TX) and added into the culture system at concentrations of 0, 5, or 10 μM 48 hours before collection of cells. The antibodies used for western blotting were as follows: anti-human p19INK4d, p18INK4c, p27KIP1, GATA1, KLF1, γ-globin (HBG), HSP70, PEBP1, ERK, AKT, RCC1, α-tubulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Santa Cruz (Dallas, TX); and anti-human p-ERK, p-AKT, HSP27, and CD44 from Cell Signaling Technology (Beverly, MA). Antibodies used for flow cytometry analysis were CD34-phycoerythrin (PE), 7AAD, glycophorin A (GPA)–PE Cy7, and α4-integrin-PE from BD Pharmingen (Franklin Lakes, NJ). Anti-human 4.1R, tropomodulin, and band 3-allophycocyanin (APC) were used previously.1,23
Cell culture
Human cord blood samples were obtained from Xiangya Hospital of Central South University or the New York Blood Center Cord Blood Program under institutional review board approval and in accordance with the Declaration of Helsinki. The detailed culture medium composition and the culture protocol have been described previously.1
Lentivirus packaging and infection
Lentivirus was packed in HEK293T cells (ATCC CRL-11268) grown in Dulbecco’s modified Eagle medium (Gibco, Grand Island, NY) with 10% fetal bovine serum (Gibco) at 37°C in 5% CO2. p19INK4d short hairpin RNAs (shRNAs) (TRCN0000233271 and TRCN0000045276) and control shRNA (luciferase shRNA [Lucif shRNA]) were obtained from Sigma (Santa Clara, CA); the detailed sequences are shown in supplemental Table 1, available on the Blood Web site. Lentivirus containing p19INK4d shRNAs or control shRNA was packaged according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA), and virus titers were measured using NIH3T3 cells (ATCC CRL-1658). Thirty million lentiviruses were used to infect 0.5 million CD34+ cells on day 2. Puromycin was added at a final concentration of 1 μg/mL to the medium on day 4 of culture to positively select for infected cells.
GATA1 rescue assay
GATA1 overexpression (GATA1-GFP, short for GATA1-IRES-GFP) and the control plasmids that have been previously described11 were packaged into lentivirus. On day 7, GATA1 overexpression or control lentivirus was infected into the erythroblasts that were previously infected with p19INK4d shRNA or corresponding control lentiviruses on day 2 of culture. Starting at day 9, the extent of terminal erythroid differentiation was monitored. Erythroblast morphology was checked starting at day 15 of culture.
siRNA transfection
GATA1 short interfering RNA (siRNA) that has been used previously24 or the control scramble siRNA was transfected into erythroblasts on day 13 of culture according to the manufacturer’s protocol (RIB&BIO, Guangzhou, China). On day 15, erythroblast morphology was checked. PEBP1 siRNA that has been previously25 described was transfected into erythroblasts on day 9 of culture. On day 11, the cells were collected for further analysis. The detailed sequences are shown in supplemental Table 1.
RNA extraction and qRT-PCR
RNA extracted from primary cultured human erythroid cells using TRIzol reagent (Invitrogen) was reverse transcripted with SuperScript first-strand synthesis system (Invitrogen) and amplified by quantitative polymerase chain reaction (qRT-PCR) with the Cycler (Bio-Rad) and appropriate primer pairs.26 The primer sequences are shown in supplemental Table 1. The data were normalized to GAPDH messenger RNA (mRNA) and calculated by the 2-ΔΔCT method.
Western blotting analysis
Whole-cell lysates from cultured cells were prepared with RIPA buffer (Thermo Fisher, Waltham, MA) in the presence of 1× protease inhibitor and 1× PhosStop cocktail (Roche, Basel, Switzerland). Protein concentration was measured using a Pierce BCA protein assay kit (Thermo Fisher). For the nuclear extraction assays, a nuclear extraction kit was used, and all procedures were performed according to the manufacturer’s instructions (Vazyme Biotech, Nanjing, China). Western blotting analysis was performed as described previously.27
Immunofluorescence
Cells were washed in 1× phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde. Fixed cells were washed with 1× PBS and incubated with HSP70, p19INK4d, or PEBP1 antibody overnight at 4°C. After washing using 1× PBS, cells were incubated in fluorescein isothiocyanate or Cy3-conjugated secondary antibody overnight at 4°C. For colocalization analysis, the cells were then incubated with a different antibody as discussed previously. Finally, the cells were incubated in 4′,6-diamidino-2-phenylindole (DAPI) for 1 hour at 37°C. Stained cells were visualized using a fluorescence microscope (Leica, Solms, Germany) or a confocal laser-scanning microscope (Nikon, Tokyo, Japan).
IP-MS
Immunoprecipitation (IP) with p19INK4d antibody or nonspecific immunoglobulin G (IgG) was performed according to the manufacturer’s protocol (Active Motif, Carlsbad, CA). The recovered products were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and further analyzed by sliver staining according to the manufacturer’s protocol (Sangon Biotech, Shanghai, China). Specific bands in sliver stained gels were excised for protein identification by liquid chromatography–tandem mass spectrometry (MS) analysis. For co-IP, IP with p19INK4d and PEBP1 antibodies or nonspecific IgG was performed according to the manufacturer’s protocol. The products were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and further analyzed by western blotting.
Flow cytometry analysis
Cells were collected every other day from day 7 to day 15 of culture. For analysis of apoptosis, 1 × 106 cells were costained with fluorescein isothiocyanate-annexin V and 7AAD according to the manufacturer’s protocol (BD Pharmingen). To monitor erythroblast differentiation, 0.25 × 106 cells in 100 μL PBS + 0.5% bovine serum albumin were stained with 7AAD, GPA-PE Cy7, α4-integrin-PE, and band 3-APC. Samples were washed twice with PBS + 0.5% bovine serum albumin before analysis. Unstained cells and PE Cy7-, PE-, and APC-conjugated IgG isotype controls were used as negative controls. For 5-bromo-2′-deoxyuridine (BrdU) staining, 1 × 106 cells were costained with BrdU and 7AAD according to the manufacturer’s protocol (BD Pharmingen). Labeled cells were analyzed with FACS Canto II (BD Pharmingen) and Cell Quest Pro software.
Cytospin preparation
A total of 1 × 105 cells in 100 μL 1× PBS were spun for 5 minutes at 400 rpm onto glass slides using the cytospin apparatus (Thermo Scientific). After air-drying for 1 minute, slides were stained with Giemsa staining solution (Sigma, Darmstadt, Germany) according to manufacturer’s instructions. Stained cells were viewed, and images were acquired with an Olympus BX51 microscope and QCapture Pro 6.0 (Tokyo, Japan).
Statistical analysis
All data are presented as the mean ± standard deviation (SD), and the results were analyzed using the SPSS 18.0 software package. Significant differences between the groups were determined by analysis of variance and Tukey range test.
Results
Expression of CDKI family members during human terminal erythroid differentiation
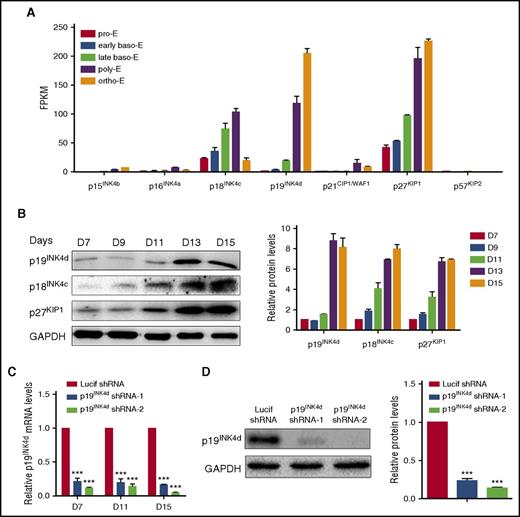
To explore the roles CDKI family members during human terminal erythroid differentiation, we first examined their expression patterns. mRNA expression levels of the CDKI members as assessed by RNA sequencing (RNA-seq) of erythroblasts cultured from cord blood CD34+ cells at distinct stages of development28 are shown in Figure 1A. Of the 7 CDKI members, the expression of 4 (p15INK4b, p16INK6a, p21CIP1/WAF1, and p57KIP2) are at low or undetectable levels. In contrast, the other 3, p18INK4c, p19INK4d, and p27KIP1, are abundantly expressed in erythroid cells and their expression is upregulated at later stages of terminal erythroid differentiation. The expression patterns of p18INK4c, p19INK4d, and p27KIP1 at the protein level were further confirmed by western blotting analysis (Figure 1B). In particular, the expression of p18INK4c and p27KIP1 increased gradually during terminal erythroid differentiation, whereas the expression of p19INK4d is dramatically upregulated after the basophilic stage. The unique expression pattern of p19INK4d probably suggests a potentially unique function. In contrast to the demonstrated roles of p18INK4c and p27KIP1 in murine terminal erythroid differentiation,13,19,22 the function of p19INK4d during either murine or human terminal erythroid differentiation has not been previously studied. To explore the role of p19INK4d during human erythropoiesis, we used a shRNA-mediated knockdown approach in human CD34+ cells. The knockdown efficiency was examined on days 7, 11, and 15 by qRT-PCR analysis. As shown in Figure 1C, an 80% to 90% knockdown efficiency was achieved with both p19INK4d shRNAs compared with control Lucif shRNA. The knockdown of p19INK4d protein was confirmed by western blotting analysis, which also showed ∼80% reduction (Figure 1D).
CDKI expression in human terminal erythroid differentiation. (A) RNA-seq data showing CDKI members’ expression (fragments per kilobase of transcript per million) at each distinct stage of human terminal erythroid differentiation. Baso-E, basophilic erythroblast; ortho-E, orthochromatic erythroblast; poly-E, polychromatic erythroblast; pro-E, proerythroblast. (B) Representative image of western blotting of p19INK4d, p18INK4c, and p27KIP1 expression in whole-cell lysates prepared from erythroblasts cultured for different times (left). Quantitative analysis of data from 3 independent experiments of protein expression levels (right). GAPDH was used as a loading control. (C) qRT-PCR results showing p19INK4d expression in erythroblasts infected with lentivirus containing control (Lucif shRNA) or p19INK4d shRNA on day (D) 7, D 11, and D 15 of culture. The results were normalized to GAPDH mRNA. (D) Representative images of western blotting showing p19INK4d expression levels in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of protein expression data from 3 independent experiments (right). The results were normalized to GAPDH protein. Statistical analysis of data of 3 independent experiments and bar plot represents mean ± SD of triplicate samples. ***P < .001.
CDKI expression in human terminal erythroid differentiation. (A) RNA-seq data showing CDKI members’ expression (fragments per kilobase of transcript per million) at each distinct stage of human terminal erythroid differentiation. Baso-E, basophilic erythroblast; ortho-E, orthochromatic erythroblast; poly-E, polychromatic erythroblast; pro-E, proerythroblast. (B) Representative image of western blotting of p19INK4d, p18INK4c, and p27KIP1 expression in whole-cell lysates prepared from erythroblasts cultured for different times (left). Quantitative analysis of data from 3 independent experiments of protein expression levels (right). GAPDH was used as a loading control. (C) qRT-PCR results showing p19INK4d expression in erythroblasts infected with lentivirus containing control (Lucif shRNA) or p19INK4d shRNA on day (D) 7, D 11, and D 15 of culture. The results were normalized to GAPDH mRNA. (D) Representative images of western blotting showing p19INK4d expression levels in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of protein expression data from 3 independent experiments (right). The results were normalized to GAPDH protein. Statistical analysis of data of 3 independent experiments and bar plot represents mean ± SD of triplicate samples. ***P < .001.
p19INK4d knockdown delays human terminal erythroid differentiation
The process of human terminal erythroid differentiation was monitored by flow cytometry using recently identified surface markers.1,29 Because the differentiation of erythroid progenitor colony-forming units of erythroid to the erythroid precursor proerythroblast is accompanied by surface expression of the erythroid specific marker GPA,30,31 we monitored GPA expression on day 7 of culture. As shown in Figure 2A, although ∼60% of cells infected with Lucif shRNA were GPA-positive, only 30% to 40% of cells infected with p19INK4d shRNA were GPA-positive, suggesting delayed differentiation from the colony-forming units of erythroid stage to proerythroblasts. Differentiation of proerythroblasts to later stage erythroblasts is characterized by increased surface expression of band 3 and decreased surface expression of α4 integrin.1 As shown in Figure 2B, the upregulation of band 3 and downregulation of α4 integrin were significantly delayed in p19INK4d knockdown cells compared with control cells, implying delayed terminal erythroid differentiation.
Effects of p19INK4don human terminal erythroid differentiation. (A) Representative images of flow cytometry analysis of GPA expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA on day 7; red numbers show statistical analysis of the GPA-positive rate from 3 independent experiments. (B) Representative images of flow cytometry analysis of band 3 and α4-integrin expression on D 7 and D 13 of GPA-positive erythroblasts infected with Lucif shRNA or p19INK4d shRNA. (C) Erythroid cell growth curves determined by manual cell counting of erythroblasts infected with Lucif shRNA or p19INK4d shRNA. (D) The cell-cycle distribution results of BrdU assay from erythroblasts infected with Lucif shRNA or p19INK4d shRNA. (E) Representative images of flow cytometry analysis of apoptosis by annexin V/7AAD staining in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (top). The red numbers indicate percentage. Quantitative analysis from 3 independent experiments is shown (bottom). Statistical analysis of the data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. *P < .05.
Effects of p19INK4don human terminal erythroid differentiation. (A) Representative images of flow cytometry analysis of GPA expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA on day 7; red numbers show statistical analysis of the GPA-positive rate from 3 independent experiments. (B) Representative images of flow cytometry analysis of band 3 and α4-integrin expression on D 7 and D 13 of GPA-positive erythroblasts infected with Lucif shRNA or p19INK4d shRNA. (C) Erythroid cell growth curves determined by manual cell counting of erythroblasts infected with Lucif shRNA or p19INK4d shRNA. (D) The cell-cycle distribution results of BrdU assay from erythroblasts infected with Lucif shRNA or p19INK4d shRNA. (E) Representative images of flow cytometry analysis of apoptosis by annexin V/7AAD staining in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (top). The red numbers indicate percentage. Quantitative analysis from 3 independent experiments is shown (bottom). Statistical analysis of the data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. *P < .05.
p19INK4d knockdown results in reduced cell growth because of increased apoptosis of late-stage erythroblasts
Cell-cycle inhibitor p19INK4d has previously been shown to inhibit cell proliferation by inducing cell-cycle arrest in a variety of cell types32,33 ; therefore, we examined whether p19INK4d knockdown affects the proliferation of erythroid cells. It was expected that downregulation of this cell-cycle inhibitor would promote cell proliferation. In marked contrast to this expectation, p19INK4d knockdown had no effect on cell growth of early-stage erythroblasts (up to day 11 of culture), but it significantly impaired the growth of late-stage erythroblasts starting from day 13 of culture (Figure 2C). To explore the mechanisms responsible for impaired cell growth, we first examined the effects of p19INK4d on cell-cycle progression by BrdU incorporation and we surprised that p19INK4d knockdown did not affect the cell-cycle progression (Figure 2D). We then examined the effects of p19INK4d knockdown on apoptosis by annexin V staining. The representative profiles of annexin V staining are shown in Figure 2E. Quantitative analysis reveals that although p19INK4d knockdown had no effect on apoptosis before day 11, there was a marked increase in apoptosis starting from day 13. Because the increase in apoptosis parallels the decrease in cell numbers, these findings imply that increased apoptosis contributes to the impaired cell growth.
p19INK4d knockdown delays human terminal erythroid differentiation by decreasing GATA1 protein levels
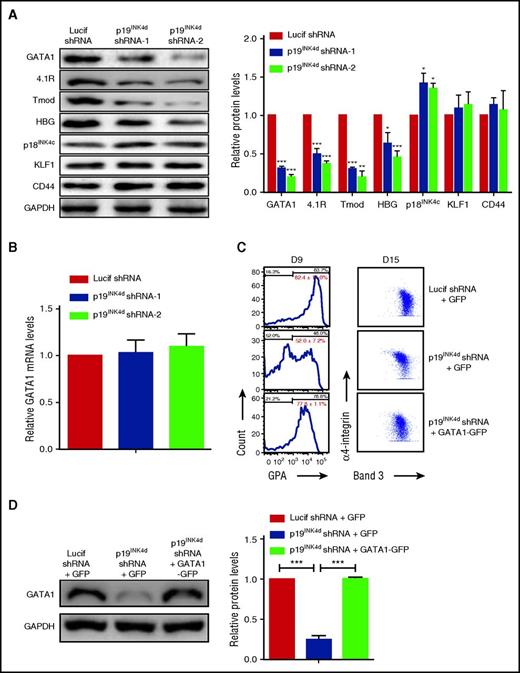
GATA1 and KLF1 are 2 major regulators of erythropoiesis.2 As an approach to explore the mechanisms for the impaired erythroid differentiation upon p19INK4d knockdown, we examined the effects of p19INK4d knockdown on the expression of these 2 transcription factors. Western blot analysis revealed that p19INK4d knockdown led to decreased levels of GATA1 protein as well as its downstream targets 4.1R, tropomodulin, and HBG.34,35 In contrast, there was no change in the expression of KLF1 and its downstream target CD4436 (Figure 3A, left). Quantitative analysis from several independent experiments revealed ∼50% to 70% reduction in expression of GATA1, 4.1R, tropomodulin, and HBG (Figure 3A, right). As shown in supplemental Figure 1, p19INK4d knockdown decreased GATA1 protein levels throughout terminal erythroid differentiation. Interestingly, p19INK4d knockdown did not affect GATA1 mRNA levels (Figure 3B), suggesting that p19INK4d posttranscriptionally modulates GATA1 protein levels. To explore whether downregulation of GATA1 is directly responsible for the impaired terminal erythroid differentiation following p19INK4d knockdown, we reintroduced GATA1-GFP in p19INK4d–transduced cells (GFP alone was used as a control). As shown in Figure 3C (left), ∼82% of control cells expressed GPA on day 9, whereas only ∼52% of p19INK4d knockdown + GFP cells expressed GPA. Importantly, close to 80% of p19INK4d knockdown + GATA1-GFP cells expressed GPA, demonstrating the rescue by GATA1 expression. Similarly, the expression pattern of band 3 vs α4-integrin was also rescued by GATA1 expression (Figure 3C, right). Reexpression of GATA1 in p19INK4d knockdown cells was confirmed by western blot analysis (Figure 3D).
p19INK4dknockdown decreased GATA1 protein expression and ectopic GATA1 expression rescues the differentiation delay. (A) Representative images of western blotting analysis showing GATA1, 4.1R, tropomodulin (Tmod), HBG, p18INK4c, KLF1, and CD44 expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (day 11 cells) (left). Quantitative analysis of protein expression data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein expression. (B) qRT-PCR analysis of GATA1 mRNA levels in erythroblasts infected with Lucif shRNA or p19INK4d shRNA. The results were normalized to GAPDH mRNA. (C) Representative data of flow cytometry analysis of erythroblasts infected with Lucif shRNA, p19INK4d shRNA, and p19INK4d shRNA combined with GATA1 overexpression. GPA expression was monitored on D 9 cells (left) and expression of band 3 and α4-integrin on D 15 cells (right). The red numbers indicate statistical analysis of GPA-positive rate from 3 independent experiments. (D) Representative images of western blotting showing GATA1 levels in whole cell lysates prepared from erythroblasts (left). Quantitative analysis of expression data from 3 independent experiments (right). GAPDH was used as a loading control. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. *P < .05, **P < .01, ***P < .001.
p19INK4dknockdown decreased GATA1 protein expression and ectopic GATA1 expression rescues the differentiation delay. (A) Representative images of western blotting analysis showing GATA1, 4.1R, tropomodulin (Tmod), HBG, p18INK4c, KLF1, and CD44 expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (day 11 cells) (left). Quantitative analysis of protein expression data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein expression. (B) qRT-PCR analysis of GATA1 mRNA levels in erythroblasts infected with Lucif shRNA or p19INK4d shRNA. The results were normalized to GAPDH mRNA. (C) Representative data of flow cytometry analysis of erythroblasts infected with Lucif shRNA, p19INK4d shRNA, and p19INK4d shRNA combined with GATA1 overexpression. GPA expression was monitored on D 9 cells (left) and expression of band 3 and α4-integrin on D 15 cells (right). The red numbers indicate statistical analysis of GPA-positive rate from 3 independent experiments. (D) Representative images of western blotting showing GATA1 levels in whole cell lysates prepared from erythroblasts (left). Quantitative analysis of expression data from 3 independent experiments (right). GAPDH was used as a loading control. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. *P < .05, **P < .01, ***P < .001.
p19INK4d knockdown leads to the generation of abnormal nuclei, which is associated with decreased GATA1 protein levels
When examining the effect of p19INK4d knockdown on cell morphology, we noted that p19INK4d knockdown led to the generation of erythroblasts with abnormal nuclei; the percentage of cells with abnormalities was ∼30% (Figure 4A). Because nuclear changes also occur in apoptotic cells,37,38 we sorted the annexin V− cells representing the nonapoptotic cell population (Figure 4B, left). Abnormal nuclei are present in nonapoptotic cells (Figure 4B, right), and this feature is not due to apoptosis. Cytospin analysis of the sorted erythroblasts at each distinct developmental stage revealed the generation of abnormal nuclei occurred specifically at the polychromatic and orthochromatic stages (Figure 4C, left). Quantitative analysis revealed that approximately 50% to 60% of polychromatic and orthochromatic erythroblasts had abnormal nuclei (Figure 4C, right). Importantly, the defects in nuclear morphology were also partially rescued by reexpression of GATA1-GFP (Figure 4D). To further examine whether there is a direct relationship between GATA1 downregulation and nuclear defects, we knocked down GATA1 in late-stage erythroblasts by siRNA (Figure 4E). As shown in Figure 4F, knockdown of GATA1 in these late-stage cells also led to generation of abnormally nucleated erythroblasts.
p19INK4dknockdown causes abnormal nuclear morphology of erythroblasts and ectopic GATA1 expression can reverse the abnormal phenotype. (A) Representative images of Lucif shRNA or p19INK4d shRNA-infected erythroblasts (day 15 cells) and quantitative analysis of cells with abnormal nuclei. (B) Flow cytometry analysis of sorted annexin V− erythroblasts (left). Representative images of sorted annexin V− erythroblasts (right). (C) Representative images of sorted erythroblasts at distinct development stages after Lucif shRNA or p19INK4d shRNA infection (left). Quantitative analysis of abnormal nuclear morphology of sorted erythroblasts at the polychromatic and orthochromatic stages from 3 independent experiments (right). (D) Representative images of erythroblasts infected with Lucif shRNA, p19INK4d shRNA, and p19INK4d shRNA combined with GATA1 overexpression (day 15 cells) (left). Quantitative analysis of nuclear morphology data from 3 independent experiments (right). (E) Representative images of western blotting showing GATA1 expression in erythroblasts transfected with GATA1 siRNA or scramble siRNA (day 15 cells) (left). Quantitative analysis of expression data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (F) Representative images of erythroblasts (day 15 cells) transfected with GATA1 siRNA or scramble siRNA (left). Quantitative analysis of nuclear morphology data from 3 independent experiments (right). The percentage of abnormally nucleated cells = counts of abnormally nucleated cells in 2000 cells/2000. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. **P < .01, ***P < .001.
p19INK4dknockdown causes abnormal nuclear morphology of erythroblasts and ectopic GATA1 expression can reverse the abnormal phenotype. (A) Representative images of Lucif shRNA or p19INK4d shRNA-infected erythroblasts (day 15 cells) and quantitative analysis of cells with abnormal nuclei. (B) Flow cytometry analysis of sorted annexin V− erythroblasts (left). Representative images of sorted annexin V− erythroblasts (right). (C) Representative images of sorted erythroblasts at distinct development stages after Lucif shRNA or p19INK4d shRNA infection (left). Quantitative analysis of abnormal nuclear morphology of sorted erythroblasts at the polychromatic and orthochromatic stages from 3 independent experiments (right). (D) Representative images of erythroblasts infected with Lucif shRNA, p19INK4d shRNA, and p19INK4d shRNA combined with GATA1 overexpression (day 15 cells) (left). Quantitative analysis of nuclear morphology data from 3 independent experiments (right). (E) Representative images of western blotting showing GATA1 expression in erythroblasts transfected with GATA1 siRNA or scramble siRNA (day 15 cells) (left). Quantitative analysis of expression data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (F) Representative images of erythroblasts (day 15 cells) transfected with GATA1 siRNA or scramble siRNA (left). Quantitative analysis of nuclear morphology data from 3 independent experiments (right). The percentage of abnormally nucleated cells = counts of abnormally nucleated cells in 2000 cells/2000. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. **P < .01, ***P < .001.
p19INK4d regulates GATA1 protein expression through the p-ERK/HSP70 pathway
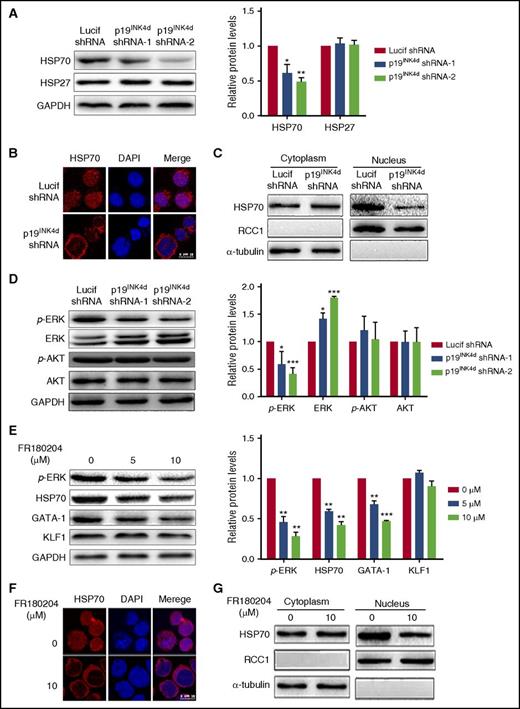
Having demonstrated the decreased GATA1 protein level upon p19INK4d knockdown, we then explored the underlying mechanisms. Because it has previously been shown that GATA1 protein levels can be regulated by HSP70 and HSP27,9,10 we examined the effects of p19INK4d knockdown on the expression levels of HSP70 and HSP27 by western blotting analysis. As shown in Figure 5A, p19INK4d knockdown dramatically decreased the HSP70 levels, but it had no significant effect on the levels of HSP27. Furthermore, because the protection of caspase 3–mediated GATA1 cleavage by HSP70 occurs in the nucleus,10 we examined the HSP70 cellular localization by immunofluorescence. Although HSP70 was predominantly localized in the nucleus of control cells, it was cytoplasmic in p19INK4d knockdown cells (Figure 5B). Western blotting analysis of nuclear and cytoplasmic fractions showed that, although the nuclear HSP70 levels decreased in knockdown cells, the cytoplasmic level was unchanged (Figure 5C). These findings strongly suggest that the reduced GATA1 protein level upon p19INK4d knockdown is due to decreased nuclear content of HSP70.
Effect of p19INK4dknockdown on HSP70 expression, localization, and ERK activity. (A) Representative images of western blotting showing HSP70 and HSP27 expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of protein expression data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (B) Representative immunofluorescence images showing HSP70 localization in erythroblasts infected with Lucif shRNA or p19INK4d shRNA. DAPI was used to stain the nucleus. (C) Western blotting analysis of nuclear and cytoplasmic fractions of HSP70. RCC1 and α-tubulin were used as nuclear and cytoplasmic markers, respectively. (D) Representative images of western blotting for p-ERK, ERK, p-AKT, and AKT in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of western blotting data from 3 independent experiments is shown (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (E) Representative images of western blotting showing p-ERK, HSP70, GATA1, and KLF1 expression level in erythroblasts treated with 0, 5, or 10 μM FR180204 (left). Quantitative analysis of western blotting data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (F) Representative immunofluorescence images showing HSP70 localization in erythroblasts treated with 0 or 10 μM FR180204. DAPI was used to stain the nucleus. (G) Western blotting analysis of nuclear and cytoplasmic fractions of HSP70 in erythroblasts from untreated and treated with 10μM FR180204. RCC1 and α-tubulin were used as nuclear and cytoplasmic markers, respectively. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. *P < .05, **P < .01, ***P < .001.
Effect of p19INK4dknockdown on HSP70 expression, localization, and ERK activity. (A) Representative images of western blotting showing HSP70 and HSP27 expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of protein expression data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (B) Representative immunofluorescence images showing HSP70 localization in erythroblasts infected with Lucif shRNA or p19INK4d shRNA. DAPI was used to stain the nucleus. (C) Western blotting analysis of nuclear and cytoplasmic fractions of HSP70. RCC1 and α-tubulin were used as nuclear and cytoplasmic markers, respectively. (D) Representative images of western blotting for p-ERK, ERK, p-AKT, and AKT in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of western blotting data from 3 independent experiments is shown (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (E) Representative images of western blotting showing p-ERK, HSP70, GATA1, and KLF1 expression level in erythroblasts treated with 0, 5, or 10 μM FR180204 (left). Quantitative analysis of western blotting data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (F) Representative immunofluorescence images showing HSP70 localization in erythroblasts treated with 0 or 10 μM FR180204. DAPI was used to stain the nucleus. (G) Western blotting analysis of nuclear and cytoplasmic fractions of HSP70 in erythroblasts from untreated and treated with 10μM FR180204. RCC1 and α-tubulin were used as nuclear and cytoplasmic markers, respectively. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. *P < .05, **P < .01, ***P < .001.
To further explore the mechanisms for the decreased nuclear HSP70, we examined ERK and AKT signal transduction pathways that have been previously shown to regulate HSP70 protein expression and localization.39,40 Figure 5D shows that, although p19INK4d knockdown had no effect AKT phosphorylation, it significantly decreased ERK phosphorylation. To further establish the causative relationship of ERK phosphorylation with HSP70 and GATA1, we treated erythroblasts with an ERK inhibitor. Figure 5E shows that similar to p19INK4d knockdown, treatment of erythroblasts with ERK inhibitor FR180204 led to decreased ERK phosphorylation, which is accompanied by the decreased HSP70 and GATA1 protein levels, although it did not affect KLF1 protein levels. Moreover, treatment of erythroblasts with ERK inhibitor FR180204 also led to decreased nuclear HSP70 (Figure 5F-G).
p19INK4d interacts with and negatively regulates PEBP1, which links p19INK4d to the ERK pathway
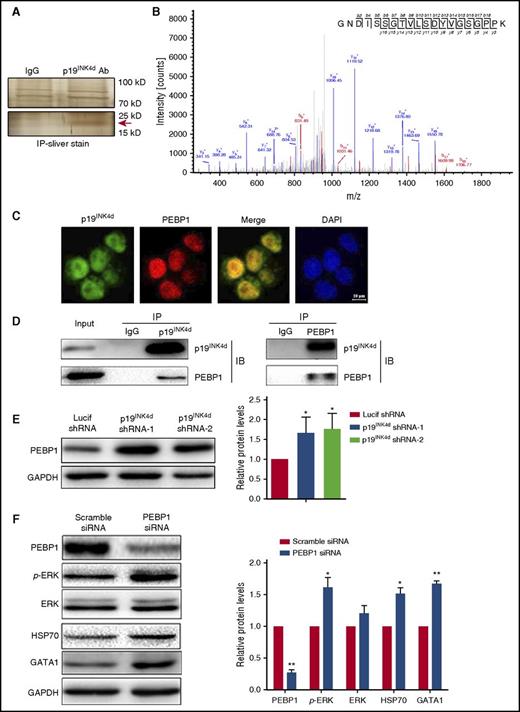
To explore the mechanism(s) by which p19INK4d knockdown induced ERK inactivation, we sought to identify p19INK4d binding partner(s) using IP followed by MS analysis. As shown in Figure 6A, anti-p19INK4d pulled down a specific band in the 15-kDa and 25-kDa range. This specific band was excised, digested with trypsin, and subjected to MS analysis for protein identification. PEBP1, which is an upstream negative regulator of ERK phosphorylation,41 was identified as a potential interaction partner of p19INK4d (Figure 6B). To examine whether PEBP1 directly associates with p19INK4d, we performed immunofluorescence staining of PEBP1 and p19INK4d. As shown in Figure 6C, p19INK4d and PEBP1 colocalize in erythroid cells. Reciprocal co-IP assays demonstrated that anti-p19INK4d antibody was able to pull down PEBP1 (Figure 6D, left), whereas the anti-PEBP1 antibody was able to pull down p19INK4d (Figure 6D, right).
p19INK4dinteracts with and negatively regulates PEBP1 and PEBP1 links p19INK4dwith the ERK pathway. (A) Representative images of silver-stained gels of IP proteins using IgG and p19INK4d antibody. (Upper) Nonspecific bands; (lower) specific band is marked with a red arrow. (B) Representative electrospray ionization-MS and MS/MS profiling of a tryptic peptide that owns higher content. The top right corner is the amino acid sequence of this peptide and the specific amino acid sequence belongs to PEBP1 through identification of proteins from the protein database. (C) Representative immunofluorescence images showing p19INK4d and PEBP1 localization in erythroblasts. DAPI was used to stain the nucleus. (D) Representative images of co-IP experiments with a p19INK4d (left) or PEBP1 antibody (right). (E) Representative images of western blotting showing PEBP1 expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of western blotting data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (F) Representative images of western blotting showing PEBP1, p-ERK, ERK, HSP70, and GATA1 levels in erythroblasts transfected with PEBP1 siRNA or scramble siRNA (left). Quantitative analysis of western blotting data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. * P < .05, ** P < .01.
p19INK4dinteracts with and negatively regulates PEBP1 and PEBP1 links p19INK4dwith the ERK pathway. (A) Representative images of silver-stained gels of IP proteins using IgG and p19INK4d antibody. (Upper) Nonspecific bands; (lower) specific band is marked with a red arrow. (B) Representative electrospray ionization-MS and MS/MS profiling of a tryptic peptide that owns higher content. The top right corner is the amino acid sequence of this peptide and the specific amino acid sequence belongs to PEBP1 through identification of proteins from the protein database. (C) Representative immunofluorescence images showing p19INK4d and PEBP1 localization in erythroblasts. DAPI was used to stain the nucleus. (D) Representative images of co-IP experiments with a p19INK4d (left) or PEBP1 antibody (right). (E) Representative images of western blotting showing PEBP1 expression in erythroblasts infected with Lucif shRNA or p19INK4d shRNA (left). Quantitative analysis of western blotting data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. (F) Representative images of western blotting showing PEBP1, p-ERK, ERK, HSP70, and GATA1 levels in erythroblasts transfected with PEBP1 siRNA or scramble siRNA (left). Quantitative analysis of western blotting data from 3 independent experiments (right). GAPDH was used as a loading control and the results were normalized to GAPDH protein. Statistical analysis of data from 3 independent experiments and bar plot represents mean ± SD of triplicate samples. * P < .05, ** P < .01.
Having demonstrated that PEBP1 is a binding partner of p19INK4d, we then examined the effects of p19INK4d knockdown on PEBP1 expression levels. As shown in Figure 6E, p19INK4d knockdown led to increased PEBP1 proteins levels. Because PEBP1 is a canonical negative regulator of ERK phosphorylation,41 the increased PEBP1 levels upon p19INK4d knockdown can account for the decreased ERK phosphorylation shown in Figure 5D. We further showed that siRNA-mediated PEBP1 knockdown led to increased ERK phosphorylation as well as increased HSP70 and GATA1 protein levels, confirming that the PEBP1-p-ERK-HSP70-GATA1 pathway is active in erythroid cells (Figure 6F).
We propose the following model (Figure 7) to integrate our findings on how p19INK4d can regulate human erythropoiesis: p19INK4d directly binds to PEBP1 and negatively regulates PEBP1 expression; p19INK4d knockdown leads to increased PEBP1 expression, which decreases ERK phosphorylation; the decreased ERK phosphorylation leads to decreased nuclear HSP70, which in turn leads to decreased GATA1 protein levels; and decreased GATA1 expression delays terminal erythroid differentiation and leads to generation of abnormally nucleated erythroblasts.
Working model of p19INK4dfunction during human terminal erythroid differentiation. The blue arrow denotes a “decreased” expression, whereas the red arrow denotes an “increased” expression. p19INK4d knockdown increases PEBP1 expression and impairs the p-ERK-HSP70-GATA1 pathway, which delays human terminal erythroid differentiation and leads to generation of abnormal nucleus.
Working model of p19INK4dfunction during human terminal erythroid differentiation. The blue arrow denotes a “decreased” expression, whereas the red arrow denotes an “increased” expression. p19INK4d knockdown increases PEBP1 expression and impairs the p-ERK-HSP70-GATA1 pathway, which delays human terminal erythroid differentiation and leads to generation of abnormal nucleus.
Discussion
Cell cycle is a fundamental cellular process. It is regulated by cyclins, CDKs, and CDKIs. Although the roles of various cell-cycle regulators have been extensively studied in many cell types,42,43 their roles in terminal erythroid differentiation remain to be fully defined. In the present study, we uncovered a previously unknown and unexpected role of a CDKI, p19INK4d, in human terminal erythropoiesis and documented that p19INK4d exerts its function via a novel pathway.
Terminal erythroid differentiation is characterized by changes in cell cycle that ultimately lead to cell-cycle exit. We found that of the 7 CDKI family members, only 3—p18INK4c, p19INK4d, and p27KIP1—are abundantly expressed in human erythroblasts and their expression is upregulated in late-stage human erythroblasts. The upregulation of p18INK4c and p27KIP1 was also observed during murine terminal erythroid differentiation.44,45 Moreover, a very recent study demonstrated that lack of KLF1 led to impaired upregulation of p18INK4c and p27KIP1 during murine terminal erythroid differentiation, which in turn led to failure in enucleation.22
In contrast to previously identified potential roles for p18INK4c and p27KIP1 in murine erythropoiesis,13,19,22 essentially nothing is known regarding the role of p19INK4d in terminal erythroid differentiation. The findings that p19INK4d knockdown impaired differentiation, inhibited cell growth, and led to increased apoptosis and generation of abnormally nucleated cells enabled the documentation of a critical role for 19INK4d during human terminal erythroid differentiation. Furthermore, the finding that cell-cycle progression is not affected following p19INK4d knockdown suggests that, in contrast to its conventional role as a cell-cycle inhibitor in many other cell types,32,33 p19INK4d has a function independent of cell cycle during human terminal erythroid differentiation. Alternatively, our finding that p18INK4c was upregulated following p19INK4d knockdown suggests that the lack of cell-cycle change of p19INK4d knockdown cells may be due to the compensatory upregulation of p18INK4c. It has been previously reported that p19INK4d knockout mice exhibited progressive hearing loss, delayed megakaryocyte maturation, and strain-specific male reproduction defects without changes in red blood cell counts in peripheral blood.46-49 The lack of anemia of p19INK4d knockout mice could be due to compensatory mechanisms that could occur in vivo but not in ex vivo culture system. Because ex vivo erythroid culture is a state of stress erythropoiesis, it will be interesting to examine whether p19INK4d knockout mice develop anemia under stress conditions. It is also interesting to note that human GATA1 and murine GATA1 do not share the caspase 3 cleavage50 ; this in turn could account for different effects of p19INK4d on human and murine erythropoiesis.
In exploring the mechanisms for phenotypic changes, we were surprised to find that GATA1 protein levels were significantly reduced upon p19INK4d knockdown. The finding that the expression levels of GATA1 downstream targets 4.1R, tropomodulin, and HBG was also decreased further supports the decreased expression of GATA1. Furthermore, it has been previously documented that GATA1 is required for the survival of terminally differentially erythroid cells.51,52 It does so by regulating the expression of antiapoptotic genes such as BCL-XL.53 It is likely that the increased apoptosis of p19INK4d-knockdown late-stage erythroblasts is due to altered expression of apoptosis-associated genes as a consequence of decreased GATA1 protein levels. Importantly, reexpression of GATA1 in p19INK4d deficient cells rescued the delayed terminal erythroid differentiation and defective nuclear morphology. Together, these findings strongly suggest that p19INK4d plays important roles in human terminal erythroid differentiation and that this is at least in part mediated by its role in regulating GATA1 protein levels.
The decrease in GATA1 protein level was not accompanied by a decrease in mRNA level, demonstrating that p19INK4d posttranscriptionally modulates GATA1 expression. In this context, our findings demonstrated that decreased GATA1 protein levels in p19INK4d knockdown cells are due to decreased nuclear HSP70. Furthermore, the findings that p19INK4d knockdown significantly decreased ERK phosphorylation and that ERK inhibitor caused similar changes in HSP70 and GATA1 levels demonstrate that p19INK4d modulates the GATA1 protein level through p-ERK-HSP70-GATA1 pathway. Finally, through a series of biochemical analyses, we identified PEBP1 as a p19INK4d-binding partner. PEBP1 is a classical Raf-1 kinase inhibitor and regulates the activity of the Raf/MEK/ERK module.41 The findings that p19INK4d knockdown increased PEBP1 expression and that PEBP1 knockdown activated ERK phosphorylation and increased HSP70/GATA1 expression indicates that PEBP1 is the link between p19INK4d and the p-ERK-HSP70-GATA1 pathway. The mechanism through which p19INK4d regulates PEBP1 is unknown. Given that p19INK4d and PEBP1 directly interact with each other, it is possible that p19INK4d may affect the stability/degradation of PEBP1.
In summary, our findings demonstrate that p19INK4d plays important roles in human terminal erythropoiesis. It does so by maintaining the GATA1 protein level via the newly identified PEBP1-p-ERK-HSP70-GATA1 pathway rather than affecting cell cycle as a canonical cell-cycle regulator. Our findings also demonstrate that posttranscriptional regulatory mechanisms can have important roles in regulating erythroid differentiation and the traditional approach of RNA-seq may miss many changes occurring at this level. Because altered HSP70 nuclear localization and GATA1 expression are associated with ineffective erythropoiesis in MDSs and thalassemias,5,6 our findings may have implications in understanding the pathogenesis of these diseases.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Natural Science Foundation of China (grants 81270576, 81470362, 81530005, and 81170459), the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (DK100810 and DK26263), the Science and Technology key project of Hunan Province (2015JC3010), and the postgraduate innovation project of Central South University of China (2015zzts095), and the Mittal students’ innovation project of Central South University of China (15MX48).
Authorship
Contribution: X.H., J.Z., Y.P., M.P., X.C., H.C., J.S., X.H., M.Y., and J.L. performed research and analyzed the data. V.G.S., C.D.H., and N.M. analyzed the data and edited the paper. X.A. and J.L. designed experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jing Liu, The State Key Laboratory of Medical Genetics & School of Life Sciences, Central South University, 110# Xiangya Rd, Changsha, 410078, China; e-mail: liujing2@sklmg.edu.cn or jingliucsu@hotmail.com; and Xiuli An, Laboratory of Membrane Biology, New York Blood Center, 310 E 67th St, New York, NY 10065; e-mail: anxl@zzu.edu.cn or xan@nybc.org.
References
Author notes
X.H. and J.Z. contributed equally to this work.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal