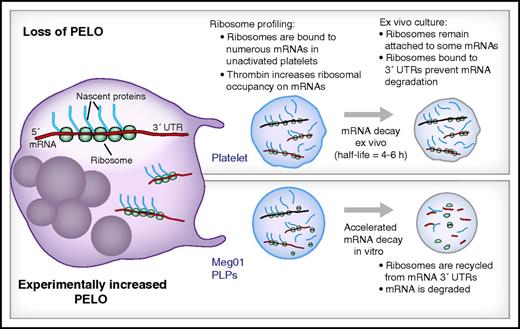
In this issue of Blood, Mills et al comprehensively examine ribosome-bound messenger RNAs (mRNAs) in resting and activated platelets and describe a role for the ribosome rescue factor Pelota (PELO) in regulating mRNA decay in platelets (see figure).1
Ribosome profiling of platelets and platelet-like particles (PLPs) demonstrates a role for PELO in mRNA decay. (Top) Megakaryocytes (left) produce platelets that are naturally devoid of PELO protein. Ribosome profiling measurements on platelets demonstrate that thousands of mRNAs are translationally active and that thrombin increases translation of many of these transcripts. Because platelets are anucleate, new mRNAs cannot be made, and their mRNA is degraded over time thus limiting translation. However, because the ribosome rescue factor PELO is naturally absent from platelets, ribosomes remain attached to the 3′ untranslated region (UTR) of platelets and slow their degradation thus potentially prolonging their availability for translation into protein. (Bottom) When PELO is transgenically increased in PLPs from a megakaryocyte cell line (Meg01), ribosomes are removed from the 3′UTR, and mRNA degradation is accelerated, potentially decreasing protein synthesis.
Ribosome profiling of platelets and platelet-like particles (PLPs) demonstrates a role for PELO in mRNA decay. (Top) Megakaryocytes (left) produce platelets that are naturally devoid of PELO protein. Ribosome profiling measurements on platelets demonstrate that thousands of mRNAs are translationally active and that thrombin increases translation of many of these transcripts. Because platelets are anucleate, new mRNAs cannot be made, and their mRNA is degraded over time thus limiting translation. However, because the ribosome rescue factor PELO is naturally absent from platelets, ribosomes remain attached to the 3′ untranslated region (UTR) of platelets and slow their degradation thus potentially prolonging their availability for translation into protein. (Bottom) When PELO is transgenically increased in PLPs from a megakaryocyte cell line (Meg01), ribosomes are removed from the 3′UTR, and mRNA degradation is accelerated, potentially decreasing protein synthesis.
Prior studies have demonstrated that nucleated megakaryocytes invest their anucleate platelet progeny with thousands of capped and poly-adenylated mRNAs2 and ribosomes.3 Platelets use this translational machinery to synthesize a subset of proteins, a function that is potentiated by platelet activation.4 Translation is higher in reticulated (ie, young) platelets, and mRNA levels and translation seem to decline as platelets age in the circulation.5 Although several targets have been identified and functionally defined,2 the repertoire of mRNAs that are translated into protein in resting and activated platelets is generally unknown. It is also unclear how mRNAs are stably expressed in circulating platelets.
To address these knowledge gaps, Mills et al screened platelets by using a sophisticated RNA sequencing (RNA-Seq)–based technique called genome-wide ribosome profiling that Ingolia et al developed in 2009.6 This technique relies on the concept that ribosomes are tightly bound to translating mRNAs and that bound sequences elude RNAse degradation whereas ribosome-free mRNAs are degraded. RNA-Seq of the ribosome-protected fragments (ie, ribosomal footprints) yields a genome-wide, single nucleotide resolution survey of translating mRNAs. In a recent report, Mills et al found that ribosomes occupy more than 6000 mRNAs in platelets.7 In their study, they found that ribosomal footprint levels generally correlate with protein levels in platelets.8 They also found that thrombin increases ribosomal occupancy on mRNAs, consistent with previous work demonstrating that bulk translation increases in activated platelets.9 Whether all or a subset of ribosomal-occupied mRNAs are actually translated into full proteins is not known and requires further validation. There is a possibility that ribosomes are stalled during translation of mRNAs, which would reveal a new mode of translational control for platelets.
The Mills et al study group previously found that ribosomes accumulate on the 3′UTR of certain transcripts in platelets and in PLPs, which have naturally low expression of the ribosomal rescue and surveillance factor PELO.7 Conversely, overexpression of PELO decreases ribosomal tracking into the 3′UTR.7 Because PELO mediates mRNA degradation when ribosomes enter the 3′UTR,10 the authors examined roles for PELO in mRNA decay in PLPs harvested from a megakar-yocytic cell line. By using RNA-Seq, they demonstrated that the half-life of mRNAs in cultured PLPs is 5.7 hours, a result that coincides with previous measurements of ex vivo mRNA decay in platelets.5 Transgenic overexpression of PELO in PLPs significantly decreased the half-life of the majority of PLP mRNAs. Given that PELO expression is low in platelets,7 these data suggest that megakaryocytes invest low levels of PELO into platelets as a mechanism for preserving mRNA levels in anucleate platelets that are incapable of transcribing new mRNA. In addition to promoting mRNA decay, the release of stalled ribosomes by PELO frees the ribosomes for additional rounds of translation. This raises the possibility that decreased expression of PELO may also be a mechanism for suppressing translation in circulating platelets, especially in resting (ie, unactivated) cells. Consistent with this possibility, reduced PELO expression in erythrocytes, another anucleate cytoplast, is associated with decreased protein synthesis.7 Because suppressed translation may accompany increased mRNA stability, the net effect of reduced PELO on individual or total protein production in platelets remains to be determined.
The fascinating articles by Mills and colleagues7 provide the first demonstration that ribosomes bind mRNAs in platelets and that ribosomal footprints are fluid, as evidenced by differential mRNA/ribosome occupancy patterns in activated platelets. Their studies also raise the possibility that different soluble agonists, adhesion to extracellular matrices, or aggregation (homotypic and heterotypic) alter platelet ribosomal occupancy patterns in a trigger-specific fashion. Other questions require further inquiry. One of these is whether results obtained in megakaryocyte-derived PLPs mimic what is observed in bona fide platelets. Studies will also be required to determine how changes in platelet protein synthesis, including targets altered by PELO, control platelet function and in vivo physiology/pathology. As has been done with mRNA profiling to identify differentially expressed mRNAs in platelets in health and disease, we anticipate that the ribosomal profiling approach pioneered by Mills and colleagues will be useful to identify the in vivo conditions and diseases that alter platelet translation and the functionally relevant differentially translated proteins.
Conflict-of-interest disclosure: The authors declare no competing financial interests.