Key Points
Only a few F9 nonsense mutations are responsive to drug-induced readthrough due to specific translation and protein structural constraints.
Reinsertion of the WT residue and gain-of-function effects account for functionally relevant readthrough.
Abstract
Drug-induced readthrough over premature stop codons (PTCs) is a potentially attractive therapy for genetic disorders, but a wide outcome variability has been observed. Through expression studies, we investigated the responsiveness to the readthrough-inducing drug geneticin of 11 rationally selected factor IX (FIX) nonsense mutations, present in 70% (324/469) of hemophilia B (HB) patients with PTCs. Among the predicted readthrough-permissive TGA variants, only 2 (p.W240X and p.R384X) responded with a remarkable rescue of FIX activity. The amounts of rescued full-length FIX protein for the p.W240X (∼9% of recombinant FIX [rFIX]–wild-type [WT]) slightly exceeded activity (5.2 ± 0.6%). FIX antigen for the p.R384X (1.9 ± 0.3%) was remarkably lower than activity (7.5 ± 0.7%). Data indicate novel specific mechanisms producing functional rescue: (1) prevalent reinsertion of the authentic residue (tryptophan), reverting the nonsense effects for the p.W240X, and (2) gain-of-function for the p.R384X, supported by the fourfold increased activity of the most probable readthrough-mediated missense variant (rFIX-R384W). For most PTCs, impaired secretion/function produced by readthrough-mediated amino acid substitutions prevented a significant functional rescue, which requires combinations of favorable FIX messenger RNA (mRNA) sequence and protein features. This rational approach, applicable to other coagulation disorders, helps with interpreting the poor response reported in the few investigated HB patients, and identifies candidate patients eligible for treatment.
Introduction
Together with other innovative approaches,1,2 the development of compounds promoting ribosome readthrough over nonsense mutations, and thus synthesis of full-length proteins, is receiving increasing attention for therapeutic purposes.3 This approach could be of particular interest for treatment of coagulation factor disorders, where even a tiny increase in functional protein levels would significantly mitigate the bleeding phenotype.4 However, the evaluation of readthrough-inducing drug efficacy in different disorders5 showed a wide outcome variability,6-9 with poor response being observed in several patients.10-13
The rescue of functional protein levels results from a complex interplay between messenger RNA and protein components: (1) the well-recognized nucleotide context of the premature stop codon (PTC), influenced by the triplet (TGA≥TAG>TAA) and the downstream nucleotide,14 and (2) the impact of amino acid changes inserted in the full-length protein during PTC mis-recognition.15 The latter, a key determinant of mutation responsiveness, has never been addressed in hemophilias.
To provide a systematic investigation by a rational selection of nonsense mutations, we evaluated quantitative and qualitative aspects of drug-induced readthrough for nonsense mutations causing hemophilia B (HB).16 In addition, to provide experimental evidence for molecular mechanisms underlying successful readthrough, we identified a few nonsense mutations, and thus patients, potentially eligible for this therapeutic approach.
Study design
Mutation selection
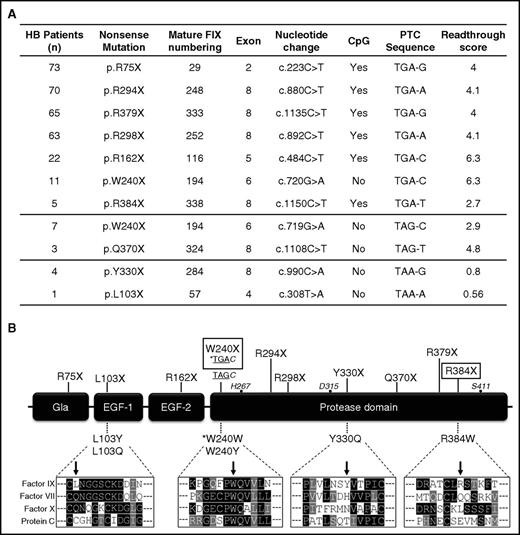
Figure 1A reports the features of the investigated nonsense mutations, chosen to include all those recurrent at CpG sites with the best-predicted susceptibility to readthrough (TGA codon)14,17,18 and 4 additional PTCs selected as controls. Altogether, these mutations have been reported in 324/469 (70%) HB patients with nonsense mutations (www.factorix.org).19
Features of F9 nonsense mutations selected for recombinant expression. (A) Features of HB-causing nonsense mutations listed according to the number of patients and type of nonsense triplet. The induced-readthrough score, ranging from 0 (unfavorable sequence) to 10 (highly favorable) was adapted from in vitro studies on G418-induced readthrough by Manuvakhova et al.14 (B) Schematic representation of FIX organization and relative position of nonsense mutations (top) and of predicted missense changes arising from readthrough (bottom). Rectangles highlight the most readthrough-responsive PTCs in our in vitro expression platform. The sequence alignments of the selected amino acid positions among the homologous FIX (NP_000124.1), FVII (NP_000122.1), FX (NP_000495.1), and protein C (NP_000303.1) are indicated below the corresponding missense variants. The asterisk indicates that readthrough over the W240X(TGAC) is expected to reinsert the authentic residue (tryptophan). The catalytic triad residues (black circles) are also numbered. EGF, epidermal growth factor–like domain.
Features of F9 nonsense mutations selected for recombinant expression. (A) Features of HB-causing nonsense mutations listed according to the number of patients and type of nonsense triplet. The induced-readthrough score, ranging from 0 (unfavorable sequence) to 10 (highly favorable) was adapted from in vitro studies on G418-induced readthrough by Manuvakhova et al.14 (B) Schematic representation of FIX organization and relative position of nonsense mutations (top) and of predicted missense changes arising from readthrough (bottom). Rectangles highlight the most readthrough-responsive PTCs in our in vitro expression platform. The sequence alignments of the selected amino acid positions among the homologous FIX (NP_000124.1), FVII (NP_000122.1), FX (NP_000495.1), and protein C (NP_000303.1) are indicated below the corresponding missense variants. The asterisk indicates that readthrough over the W240X(TGAC) is expected to reinsert the authentic residue (tryptophan). The catalytic triad residues (black circles) are also numbered. EGF, epidermal growth factor–like domain.
Expression vectors and transfection
Creation of expression vectors for recombinant factor IX (rFIX) nonsense and missense variants (Figure 1B) by site-directed mutagenesis of FIX complementary DNA cloned in pCMV5, and transient transfection of human embryonic kidney 293 (HEK293) cells were performed as described.8 Sequences of mutagenic primers are available on request. Transfected cells were incubated with an optimized geneticin concentration (100 µg/mL).6,9 Media and cell lysates were collected 48 hours later.
Evaluation of rFIX protein
Activity of rFIX variants was evaluated by activated partial thromboplastin time (aPTT)-based assays20 and a commercially available chromogenic assay (Biophen; Aniara Diagnostica, West Chester, OH). Coagulation times or optical density values from serial dilutions of rFIX–wild-type (WT) were used as reference.
Secreted rFIX levels were evaluated by enzyme-linked immunosorbent assay (ELISA) (Affinity Biologicals, Ancaster, ON, Canada) using plasma-derived human FIX (Haematologic Technologies, Essex Junction, VT) as reference.
FIX isoforms were evaluated by western blotting through polyclonal goat anti-human FIX (APGAFIX; Affinity Biologicals) and anti-goat horseradish peroxidase-conjugated (A50-101P; Bethyl Laboratories, Montgomery, TX) antibodies. Blotting images were analyzed by Image Laboratory Software version 4.0 (Bio-Rad, Hercules, CA).
Results and discussion
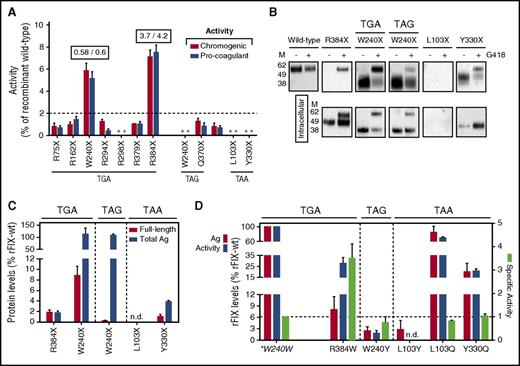
In this study, prompted by the wide outcome variability observed in patients, we evaluated the responsiveness to readthrough-inducing drugs of the most recurrent HB-causing nonsense mutations (Figure 1A) by exploiting an in vitro expression platform. We therefore expressed the rFIX nonsense variants (Figure 1B) in HEK293 and challenged them with a well-established readthrough-inducing agent, geneticin (G418), to assess the key outcome, which is the increase in FIX activity levels over the undetectable basal levels. As shown in Figure 2A, G418 elicited a poor rescue, if any, for the large majority of mutations, considering as threshold for its potential therapeutic meaning levels >2% of rFIX-WT. Remarkably, the rFIX-R384X (TGAT; chromogenic 7.1 ± 0.6%; pro-coagulant 7.5 ± 0.7%) and the rFIX-W240X(TGAC; 5.6 ± 0.7% and 5.2 ± 0.6%) displayed a robust rescue. By comparison, the rFIX-W240X(TAGC), which is the topologically overlapping nonsense variant at the W240X PTC differing in the nonsense triplet, did not appreciably respond to G418.
Evaluation of drug-induced readthrough over F9 nonsense mutations. (A) FIX activity levels in medium from cells expressing the rFIX nonsense variants upon treatment with G418, evaluated by chromogenic (red bars) and aPTT-based (blue bars) assays. Numbers above the bars report the activity/antigen ratio of the most responsive variants. Nonsense variants are indicated by amino acid numbering and grouped according to the 3 nonsense triplets (theoretical readthrough susceptibility, TGA≥TAG>TAA). The dotted line represents the selected threshold of 2%. The activity in medium from untreated cells was undetectable for all variants and the undetectable activity after treatment is indicated by asterisks. (B) Western blotting analysis on secreted (top) and intracellular (bottom) rFIX proteins transiently expressed from HEK293 cells untreated (−) or treated (+) with G418. The W240X(TGAC) and R384X stop codons, producing the highest rescue, are compared with PTCs displaying barely detectable (W240X and Y330X) or undetectable (L103X) readthrough. For the W240X PTCs, both nonsense triplets are indicated. The images are representative of at least 3 independent experiments. (C) Secreted full-length (red bars) and total (blue bars) rFIX levels after G418 treatment. Full-length rFIX levels were calculated by densitometric analysis of western blots shown in (B) and total antigen by ELISA. (D) Antigen (red bars), pro-coagulant activity (blue bars) levels, and activity/antigen ratio (specific activity, light green bars) of the most probable rFIX missense variants arising from misrecognition of TGA (R384W), TAG (W240Y), and TAA (L103Y/L103Q and Y330Q) stop codons. Readthrough over the W240X(TGAC) PTC is predicted to reintroduce the authentic amino acid (tryptophan, *W240W; Figure 1B). The dashed-line indicates the specific activity of WT rFIX. Results in (A,C-D) are reported as mean ± standard deviation from at least 3 independent experiments. Ag, antigen; M, molecular weight marker; n.d., not detectable.
Evaluation of drug-induced readthrough over F9 nonsense mutations. (A) FIX activity levels in medium from cells expressing the rFIX nonsense variants upon treatment with G418, evaluated by chromogenic (red bars) and aPTT-based (blue bars) assays. Numbers above the bars report the activity/antigen ratio of the most responsive variants. Nonsense variants are indicated by amino acid numbering and grouped according to the 3 nonsense triplets (theoretical readthrough susceptibility, TGA≥TAG>TAA). The dotted line represents the selected threshold of 2%. The activity in medium from untreated cells was undetectable for all variants and the undetectable activity after treatment is indicated by asterisks. (B) Western blotting analysis on secreted (top) and intracellular (bottom) rFIX proteins transiently expressed from HEK293 cells untreated (−) or treated (+) with G418. The W240X(TGAC) and R384X stop codons, producing the highest rescue, are compared with PTCs displaying barely detectable (W240X and Y330X) or undetectable (L103X) readthrough. For the W240X PTCs, both nonsense triplets are indicated. The images are representative of at least 3 independent experiments. (C) Secreted full-length (red bars) and total (blue bars) rFIX levels after G418 treatment. Full-length rFIX levels were calculated by densitometric analysis of western blots shown in (B) and total antigen by ELISA. (D) Antigen (red bars), pro-coagulant activity (blue bars) levels, and activity/antigen ratio (specific activity, light green bars) of the most probable rFIX missense variants arising from misrecognition of TGA (R384W), TAG (W240Y), and TAA (L103Y/L103Q and Y330Q) stop codons. Readthrough over the W240X(TGAC) PTC is predicted to reintroduce the authentic amino acid (tryptophan, *W240W; Figure 1B). The dashed-line indicates the specific activity of WT rFIX. Results in (A,C-D) are reported as mean ± standard deviation from at least 3 independent experiments. Ag, antigen; M, molecular weight marker; n.d., not detectable.
To dissect the molecular bases of these observations, we investigated the intracellular and secreted FIX species by western blotting, and expressed the most probable missense variants expected by readthrough.15 The interpretation of results illustrates different scenarios.
Expression of the p.R384X mutation showed both full-length and truncated forms at the intracellular level (Figure 2B). However, only full-length FIX was secreted in medium, which accounted for 1.9 ± 0.3% of rFIX-WT (Figure 2C). This value was much lower than the measured activity (7.5 ± 0.7%), thus pointing out hyperactive features, with an estimated fourfold increased activity/antigen ratio. Consistently, the most probable missense variant (rFIX-R384W) arising from readthrough (Figures 1B and 2D), albeit with reduced secretion (8.0 ± 3.3%), revealed an increased specific activity (3.7 ± 0.5-fold). Similar to the thrombophilic R384L substitution (FIX Padua),21-23 the R384W substitution confers gain-of-function features, thus magnifying the drug effects that are higher than those expected from the readthrough score (Figure 1A).
Intriguingly, the p.R384X nonsense mutation, an example of recurrent C>T change at a CpG site,24 has been detected in only 5 HB patients (2 moderate), a number much lower than that of the other TGA PTCs (n = 22 to 73; Figure 1A), which stimulates further genetic and epigenetic investigation in vivo.
For the rFIX-W240X(TGAC), the amount of full-length rFIX protein evaluated by western blotting (Figure 2B-C) was much higher (8.8 ± 1.8% of rFIX-WT) than for the rFIX-W240X(TAGC) variant (0.25 ± 0.1%), despite the indistinguishable secreted levels measured by ELISA (Figure 2C). This is attributable to counterbalancing amounts of truncated molecules. Intriguingly, the sequence context of the favorable W240X(TGAC) PTC predicts, as the most probable event during readthrough, the reinsertion of tryptophan (82% of events),15 which would lead to the WT FIX molecule. Although with much lower frequency, cysteine (14%) and arginine (4%) could also be introduced, but the p.W240C25 and p.W240R26 missense changes, found in severe HB patients, are associated with barely detectable FIX levels. Consistently, the rFIX-W240Y missense variant, predicted to arise from readthrough over the unfavorable W240X(TAGC) PTC, was secreted with very poor efficiency (2.5 ± 1% of rFIX-WT; Figure 2D). These coherent findings support that the highly conserved tryptophan (Figure 1B) is the sole residue compatible with the functional improvement observed in response to G418 for W240X(TGAC). For this variant, the apparently reduced activity/antigen ratio (0.6 ± 0.1) is very likely produced by the secretion of truncated molecules, which is also abundant at the intracellular level (Figure 2B).
Altogether, these data corroborate the remarkable nucleotide sequence-related differences in susceptibility to drug-induced readthrough, and anticipates unexpected differences in the response of patients with apparently identical nonsense mutations.
The balance between mechanisms accounting for poor response of the W240X(TAGC) also pertain to the other investigated unfavorable nonsense mutations, as in the paradigmatic examples of p.L103X(TAAA) and p.Y330X(TAAG). Expression of the predicted missense variants indicates that these moderately conserved positions (Figure 1B) are functionally tolerant to amino acid changes (Figure 2D), but their biosynthesis is prevented by inefficient, predicted (Figure 1A), and observed (Figure 2A-C), ribosome readthrough.
In conclusion, the extensive investigation of readthrough over PTCs reported in the vast majority of HB patients:
Provides novel elements for interpretation of the ample variability in the response to readthrough-inducing drugs. Our data explain the pioneering results previously obtained with aminoglycosides in HB patients, showing a small magnitude hemostatic response for the p.R298X and p.R379X mutations,10 and may interpret the high responder p.R384X HB mouse model7
Indicates for the first time in HB that readthrough-favorable messenger RNA sequence contexts are heavily selected by protein constraints. In FIX, and very likely in other diseases caused by secreted enzyme defects, favorable mechanisms such as reinsertion of the WT residue or rare gain-of-function effects may play a key role in achieving a therapeutically relevant treatment output
Proposes a rational approach contributing to identifying nonsense mutations, and thus patients, potentially eligible for therapeutic options based on readthrough
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was financially supported by Pfizer EUROASPIRE (projects WI193137 and WI199905) (M.P. and A.B., respectively), and Telethon-Italy (GGP14190) (M.P.).
Authorship
Contribution: A.B. and M.F. expressed and characterized recombinant variants; M.C. created expression plasmids; R.M. performed aPTT-based assays; G.C. analyzed the data and carefully revised the manuscript; M.P., A.B., and F.B. conceived the study and designed research, analyzed and interpreted data, and wrote the manuscript; and all authors revised and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alessio Branchini, Department of Life Sciences and Biotechnology, University of Ferrara, via Fossato di Mortara 74, 44121 Ferrara, Italy; e-mail: brnlss@unife.it.
References
Author notes
A.B. and M.F. contributed equally to this study.