In this issue of Blood, Nakata et al demonstrate that hematopoietic-specific expression of a mutation in the E3 ubiquitin ligase Casitas B cell lymphoma (CBL) results in a disorder that recapitulates the key features of human chronic myelomonocytic leukemia (CMML).1 One of the challenges currently facing researchers studying CMML is the scarcity of faithful model systems to study disease pathogenesis and therapeutic response. Given the lack of curative therapy for most patients with CMML, there is a pressing need to develop improved preclinical models to inform mechanistic studies and drug development. Although genetically engineered murine models exist for the 3 most commonly mutated genes in CMML, SRSF2, TET2, and ASXL1,2 none of these models recapitulates the entire phenotypic spectrum of human CMML, including chronic elevation in monocytes, multilineage dysplasia, hypersensitivity to cytokines, and susceptibility to transformation to acute myeloid leukemia (AML).
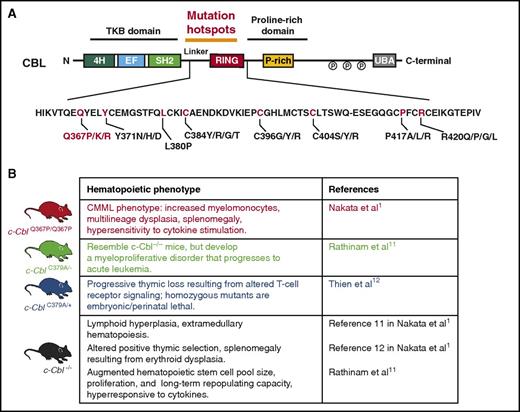
CBL mutations and currently published mouse models of mutant Cbl. (A) Protein domains of CBL with location of mutation hotspots in the linker and RING domain regions of the protein. In red are the recurrently mutated amino acid residues in myeloid neoplasms, with the Q367P mutation highlighted. (B) Published mouse models of Cbl mutations, including the newly described CblQ367P/Q367P model by Nakata et al.
CBL mutations and currently published mouse models of mutant Cbl. (A) Protein domains of CBL with location of mutation hotspots in the linker and RING domain regions of the protein. In red are the recurrently mutated amino acid residues in myeloid neoplasms, with the Q367P mutation highlighted. (B) Published mouse models of Cbl mutations, including the newly described CblQ367P/Q367P model by Nakata et al.
CBL is commonly mutated across myeloid malignancies, with the greatest enrichment of mutations occurring in CMML and juvenile myelomonocytic leukemia. In fact, CBL mutations were, in part, discovered by searching for the genetic basis for acquired uniparental disomy on chromosome 11q in patients with CMML. This led to the identification of homozygous point mutations in CBL resulting from acquired uniparental disomy. However, prior efforts to genetically manipulate Cbl in vivo with Cbl−/− or Cbl-mutant constitutive murine models3-5 resulted in mice with either lymphoid and/or myeloproliferative disorders with spontaneous progression to AML without strong features of CMML (see figure). Now, using a conditional knock-in mouse model approach, the authors were able to inducibly express a commonly occurring mutation in Cbl (the CblQ367P mutation) in a homozygous state to mimic the most common genetic configuration in CBL-mutant patients with CMML.6,7 CblQ367P/Q367P mice developed classic hallmarks of CMML, including sustained elevation and proliferation of myelomonocytes, dysplastic hematopoiesis, and splenomegaly, as well as hyperactivation of prosurvival signaling pathways (JAK-STAT and PI3K-AKT). Interestingly, the phenotypic effects of the CblQ367P mutation were most evident and severe in the homozygous state, consistent with the frequent occurrence of this mutation in the homozygous state in patients. Moreover, the authors demonstrate that the CMML disease emanates from long-term hematopoietic stem cells in transplantation assays. Although these results are fascinating and informative, it will also be important to determine whether the disease-initiating capability conferred by the CblQ367P/Q367P alteration is restricted to the hematopoietic stem cell compartment vs also being present in more committed myeloid progenitors such as common myeloid progenitors or granulocyte-macrophage progenitors. This is an important question, as functional evaluation of the cell of origin of CMML from patients has not been performed yet because of the lack of robust in vivo CMML models. Given reports of patients with germline mutations in CBL, it will also be interesting to assess the effects of expressing the CblQ367P mutation in early embryonic or early hematopoietic development, using Cre-recombinases such as EIIa-Cre or Vav-Cre.
To identify potential genetic alterations that would transform the CblQ367P/Q367P-induced CMML into AML, the authors performed unbiased mutagenesis screens. Using retroviral insertion screen approaches, they determined that Evi1 overexpression collaborates with the CblQ367P mutation to drive transformation to AML. Given the frequency of SRSF2, TET2, and ASXL1 mutations in CMML, however, it would be a fascinating next step to investigate the effect of combinatorial expression of the CblQ637P mutation with Srsf2, Tet2, or Asxl1 mutant mice. This would be especially interesting, as genetically engineered murine models of myelodysplastic syndromes are also quite limited, and it is possible that these compound mutant Cbl mice may generate models with more severe dysplastic myelodysplastic syndrome features with increased risk for leukemic transformation.
CBL mutations are often mutually exclusive with NRAS and/or KRAS activating mutations in myeloid neoplasms,2,8 suggesting the possibility of shared downstream targets and pathways perturbed by these mutations. Using transcriptomic profiling, the authors noted that CblQ637P/Q367P hematopoietic stem cells expressed higher levels of the putative oncogene Gem, similar to findings from a previous report from NRasG12D hematopoietic stem cells.9 Moreover, they confirmed that enforced expression of Gem alone in normal bone marrow is sufficient to drive CMML in vivo. It would therefore be interesting to next evaluate exactly how mutant CBL upregulates Gem, and whether GEM itself might serve as a potential therapeutic target for malignancies driven by activating RAS and CBL mutations.
One of the hopes of developing novel disease models is to effectively use them as a preclinical platform for exploring novel therapeutic approaches. Although the CblQ367P/Q367P mice represent a faithful CMML model, CBL mutations in patients with CMML may not always be present in the founding clones, as they are in the genetically engineered murine model discussed here, and results from preclinical trials using these reagents needs to be interpreted with this in mind. Nonetheless, the authors used this model to show potential promising efficacy of combined PI3K and JAK2 inhibition, a combinatorial therapeutic approach in need of further study for chronic myeloid malignancies.
As mentioned earlier, the most well-characterized role of the enzymatic activity of mutant CBL is ubiquitinylating receptor tyrosine kinases for proteasome degradation as part of the negative feedback mechanism after cytokine stimulation.10 This model now provides a platform to identify the effects of CBL mutations on the proteome in specific subsets of primary cells, including rare cell types such as stem cells. This effort, in turn, may identify novel downstream substrates of mutant CBL that mediate its leukemogenic effects and might serve as novel therapeutic targets themselves. Overall, the model generated by Nakata et al represents one of the most faithful genetically engineered murine models of human CMML to date, as well as a valuable reagent to expand our understanding of CMML pathogenesis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.