In this issue of Blood, Rossi et al demonstrate that the “liquid biopsy” has a future in the management of patients with lymphoma.1
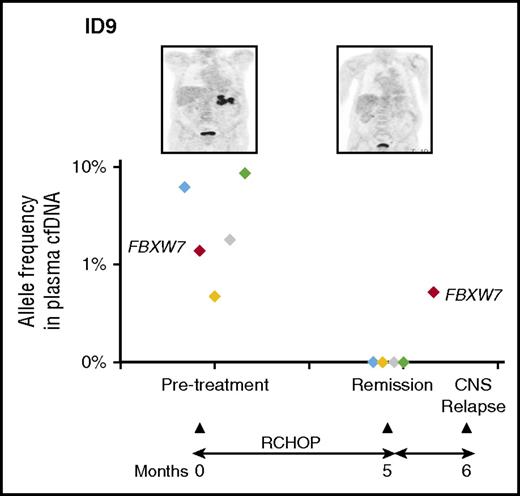
Mutational profile from cfDNA in this patient (ID9) revealed a series of mutations in the pretreatment sample. Interestingly, the FBXW7 mutation was not detected in the corresponding tumor biopsy. This mutation persisted despite clearance of other mutations from cfDNA and clinical remission. The patient relapsed in the CNS and the mutation was detected at a low level in cells from the cerebrospinal fluid. See Figure 7 in the article by Rossi et al that begins on page 1947.
Mutational profile from cfDNA in this patient (ID9) revealed a series of mutations in the pretreatment sample. Interestingly, the FBXW7 mutation was not detected in the corresponding tumor biopsy. This mutation persisted despite clearance of other mutations from cfDNA and clinical remission. The patient relapsed in the CNS and the mutation was detected at a low level in cells from the cerebrospinal fluid. See Figure 7 in the article by Rossi et al that begins on page 1947.
Circulating tumor DNA (ctDNA) is the component of cell-free DNA (cfDNA) released into serum from tumor cells as a result of apoptosis, necrosis, and secretion. Over the last few years, researchers have begun to explore the possibility of diagnosis and monitoring of solid tumors using cfDNA.
In this proof-of-principle study, plasma samples were collected pretreatment, during rituximab-cyclophosphamide-doxorubicin-vincristine-prednisone (R-CHOP) immunochemotherapy, after therapy, and at progression from 30 patients with de novo diffuse large B-cell lymphoma (DLBCL). cfDNA was isolated and subjected to next generation sequencing (NGS) using a target capture strategy designed to test for mutations in 59 lymphoma-associated genes to a depth of at least 1000 times coverage in 80% of gene targets. Germ line DNA from patient granulocytes was also evaluated to ensure mutations detected were not spurious. At diagnosis, mutations were found in 67% of patients’ serum. Among the most frequently mutated genes were KMT2D, TP53, and CREBP (20%-30% of cases), PIM1, TNFAIP3, EZH2, STAT6, TBL1XR1, B2M, BCL2, CARD11, CCND3, STAT6, and FBXW7 (10%-20% of cases). Importantly, analysis of 6 healthy donors showed no mutations.
To confirm that these mutations were also in the lymphoma tissue, 17 paired frozen samples were also analyzed. Seventy-nine percent of mutations seen in genomic tumor-derived DNA were found in the paired cfDNA. Limiting the tissue mutations to those with >20% variant allele frequency showed 95% of the mutations could be found in cfDNA. Of note, cfDNA samples from 2 of these 17 cases lacked detectable mutations. Thus, a minority of patients did not have abnormalities at diagnosis in cfDNA. Although 1 patient had stage 1 disease, the other had stage 4 disease suggesting that tumor burden is not the only factor. With regard to specificity, there were 16 mutations detected in the cfDNA samples that were not detected in tissue. Given the over 15 million potential nonsynonymous variants in the sequenced regions, this represents a very low potential false-positive rate (specificity of over 99.9%), assuming error is randomly distributed. An alternative consideration, with precedent in other systems, is that these mutations are not false positives but derive from a subclone present at a site other than that which was biopsied. This interpretation is supported by an example case (see figure) in which a FBXW7 mutation seen in the pretreatment cfDNA sample was not found in the diagnostic biopsy tissue. However, it was present in the central nervous system (CNS) relapse sample and in a remission cfDNA sample before clinical relapse. Finally, cfDNA samples collected during therapy and in follow-up showed clearance of mutations in responding patients but not in refractory patients. New mutations were detected in refractory/relapsing patient ctDNA, potentially providing insight into clonal evolution, possible resistance mechanisms, and therapeutic targets.
This is not the first study examining cfDNA in DLBCL patients and, in aggregate, the evidence is mounting that cfDNA will have a role in diagnosis and management of patients with lymphoma. ctDNA is present at diagnosis in most if not all patients with DLBCL.2,3 Two NGS strategies for analyzing ctDNA have been used. One has focused on immunoglobulin receptor IGH VDJ and IGK VJ gene rearrangement, a unique clonal marker for an individual patient’s B-cell lymphoma. By sequencing a patient’s lymphoma tissue to define the lymphoma-specific IGH VDJ and IGK VJ sequences (clonotype) and then analyzing cfDNA to search for the lymphoma clonotype,4 investigators have shown feasibility and promising results. In a small series of DLBCL patients, ctDNA was superior to markers of disease burden such as lactate dehydrogenase in detecting active disease, correlated with mean tumor volume by positron emission tomography/computed tomography, and could detect disease at, or even before, relapse.5 A larger series showed that a clonotype could be defined from biopsy tissues in 94 of 109 patients (86%) and from pretreatment serum from 54 of 86 samples (63%). Analysis of interim and surveillance serum samples showed that interim positivity predicted time to progression, and surveillance sample status had high positive and negative predictive values for recurrence, with identification of ctDNA up to 200 months before clinical relapse.6 The other strategy (explored by Rossi et al) involves sequencing a selected set of genes known to be recurrently mutated in DLBCL, with or without simultaneous assessment for clonotype and common gene fusions.3,7 This approach appears to be applicable in the great majority of cases and can provide rich data on the lymphoma mutational profile, clonal evolution, and resistance mechanisms. Furthermore, it may provide guidance for further therapy. Studies such as these suggest that this strategy also provides prognostic information, correlates with relapse, detects minimal residual disease prior to relapse, and outperforms clonotype analysis alone.3 Of course, there is greater complexity in assay design, bioinformatic algorithms, and interpretation but preanalytical, technical, and computational advances will overcome these challenges.8
There is a pressing need for better methods to assess response, monitor, and identify resistance mechanisms and therapeutic targets in order to bring personalized medicine to fruition for the management of patients with DLBCL and other lymphomas. Feasibility and initial performance characteristics of cfDNA analysis are being established by several groups and provide tantalizing glimpses into the future.1,3,5-7 Incorporation of these assays into clinical trials to define the role of cfDNA analysis in staging, risk stratification, interim response assessment, monitoring, and therapy selection will be required in order to use the liquid biopsy for routine care. The vast amount and rich content of this information, all derived from a few milliliters of serum, will transform the way in which we diagnose and manage patients with lymphoma.
Conflict-of-interest disclosure: E.D.H. has been a consultant for Seattle Genetics, HTG Molecular Diagnostics, and Alexion within the last year and receives research support from Eli Lilly, AbbVie, and Cellerant Therapeutics.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal