In this issue of Blood, Kim et al provide evidence that the t(9:22) translocation (ie, Philadelphia chromosome, or Ph), coding for the BCR/ABL gene rearrangement, is not always the first mutation acquired in chronic myeloid leukemia (CML).1 These results are consistent with prior smaller studies and indicate that CML can sometimes arise from a preexisting Ph-negative (Ph−) hematopoietic clone already harboring additional gene mutations. This work also adds to the growing body of evidence suggesting that BCR/ABL expression alone may not be sufficient to induce CML.
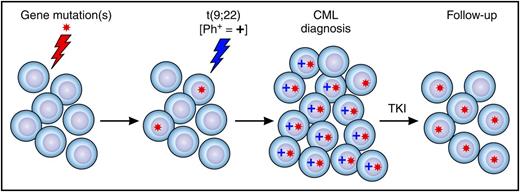
Preleukemic cells in CML and clonal hematopoiesis after TKI therapy. A hematopoietic stem and progenitor cell acquires a gene mutation or mutations and clonally expands (ie, becomes a preleukemia clone). A preleukemia cell gains a Ph chromosome and expands, and clinical CML develops. After treatment with a TKI, Ph+ cells are eradicated, and residual mutant Ph− preleukemic cells expand and contribute to clonal hematopoiesis.
Preleukemic cells in CML and clonal hematopoiesis after TKI therapy. A hematopoietic stem and progenitor cell acquires a gene mutation or mutations and clonally expands (ie, becomes a preleukemia clone). A preleukemia cell gains a Ph chromosome and expands, and clinical CML develops. After treatment with a TKI, Ph+ cells are eradicated, and residual mutant Ph− preleukemic cells expand and contribute to clonal hematopoiesis.
Thirty-five years ago, Fialkow et al first suggested that the BCR/ABL translocation may not always be the initiating mutation in CML. The authors reported that both Ph− and Ph+ cells from patients with CML can be clonal, based on the pattern of X-chromosome inactivation.2 This provided evidence that in some patients, Ph may arise in a preexisting Ph− clonal population, akin to clonal hematopoiesis of indeterminate potential (CHIP).3 Decades later, Schmidt et al reached similar conclusions by sequencing 25 genes in blood cells from 29 patients with CML.4 Ph− and Ph+ cells were isolated from patients with CML, including a subset of patients that appeared to develop clonal cytogenetic abnormalities in their Ph− cells during treatment with a tyrosine kinase inhibitor (TKI). Several patients had the same gene mutation present in both their Ph− and Ph+ cells, including 1 patient with a DNMT3A mutation in both cell types. These data provided further evidence that Ph can arise in an expanded Ph− clone carrying a CHIP-related gene mutation (ie, a “preleukemic” clone with CHIP).
Several lines of evidence from mouse and human studies also suggest that BCR/ABL expression alone may not be sufficient for disease induction. Although overexpression of BCR/ABL using a retrovirus can induce a myeloproliferative disease in mice, a knock-in model expressing physiologic levels of human BCR/ABL p210 from the mouse Bcr locus did not induce leukemia.5 In addition, there are marked strain differences (DBA/2 vs C57BL/6) in disease induction after BCR/ABL retroviral overexpression,6 suggesting that additional inherited polymorphisms influence the ability of BCR/ABL to induce disease. Finally, several groups have identified individuals who express BCR/ABL without evidence of CML.7 Collectively, the data indicate that additional genetic mutations or variants may cooperate with BCR/ABL to induce CML. Identifying mutations that co-occur with Ph in CML was explored in the current study.
To investigate the acquisition, persistence, and clearance of gene mutations in CML, Kim et al sequenced 92 myeloid malignancy–associated genes in hematopoietic cells from 100 patients with CML at diagnosis (all harboring BCR/ABL) and at follow-up after treatment with a TKI.1 The authors used T cells isolated from diagnostic samples as control tissue and identified 37 patients with gene mutations present in at least 1 timepoint. Chromatin modification and DNA methylation genes (epigenetic modifiers) were the most common category of mutant genes in the cohort at diagnosis, and spliceosome gene mutations were infrequent. Using 2013 European LeukemiaNet response criteria, patients were classified as TKI responsive, resistant, or progressed on therapy. The fraction of patients with at least 1 mutation was highest among the group that progressed on therapy, intermediate among resistant patients, and lowest in patients who responded to TKIs. As expected, ABL1 kinase domain mutations were specific to TKI nonresponsive patients.
Overall, up to 10% of patients had persistence of gene mutations in their follow-up samples after responding to TKI therapy, some without detectable mutations in their diagnostic T cells. This indicates that the t(9;22) translocation was not the first mutation in their cells (see figure). In addition, nearly all the bone marrow cells were clonal in several of these patients at follow-up, even when a 4.0 log reduction in BCR/ABL levels was achieved. These results indicate that Ph− clonal hematopoiesis can occur in patients with CML after TKI therapy. The majority of patients with CML with evidence of clonal hematopoiesis after TKI treatment had a mutation in a gene recently identified in healthy individuals with CHIP, including mutations in DNMT3A, TET2, ASXL1, BCOR, and CREBBP.3 These mutations were present in both diagnosis and follow-up samples. Collectively, the data suggest that residual “preleukemic” mutant cells lacking BCR/ABL can expand and contribute to clonal hematopoiesis after successful TKI treatment, a finding similar to that recently described for acute myeloid leukemia.8
Although the results are provocative, there are several technical issues to consider. It would be interesting to know whether BCR/ABL was detected in the T-cell fraction. Early reports suggested that BCR/ABL may occur in T cells,9 a finding that could alter the interpretation of whether a patient had preleukemic mutations in Ph− cells vs mutations in Ph+ stem cells that were capable of producing T cells. Because T cells were used as matched “normal” samples for variant calling, mutations present at high levels in T cells may be missed in diagnostic or follow-up samples (ie, false-negative mutations). Therefore, the authors used several approaches to identify preleukemic mutations present in T cells, including classifying a variant as somatic in a sample, regardless of the T-cell variant allele fraction (VAF) for that patient, if the same variant was identified as somatic in at least 1 other patient, using their matched T cells. Two variants (TET2 p.F868L and ASXL1 p.630_637del) were identified using this approach, and all 5 patients with mutation pattern 4 or 5 harbored one of the variants. These patients had similar or higher VAFs in their T cells compared with diagnostic or follow-up samples. The authors concluded that these 5 patients likely had preleukemic mutations. However, the TET2 p.F868L variant occurs at 1.5% in the East Asian population,10 raising the possibility that this variant calling approach may have limitations that could influence the results of the study.
The authors note that patients with CML with clonal hematopoiesis responded well to TKI therapy and maintained durable responses. Larger studies with longer follow-up will be needed to confirm this finding, and possibly to address whether clonal hematopoiesis is prognostic for patients with CML who discontinue TKI therapy. This study was not designed to determine whether CML arises from antecedent CHIP in some patients, but the data strongly suggest this does occur. This study also provides additional evidence suggesting BCR/ABL may not be sufficient on its own to induce CML in all patients. Certain antecedent mutations associated with CHIP may create a state of “fitness” that makes it more likely that BCR/ABL will move a cell forward to clinically recognizable CML. Identifying the full complement of genetic events that cooperate with BCR/ABL, using more unbiased sequencing approaches (eg, exome or whole genome sequencing), could nominate novel genetic dependencies that cooperate with BCR/ABL to induce CML.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal