Key Points
XLF belongs to the NHEJ ligation complex and has a dual role in DNA double-strand break repair and V(D)J recombination.
XLF is involved in N-nucleotide addition, and thereby contributes to junctional diversity of the antigen receptors.
Abstract
Repair of DNA double-strand breaks (DSBs) by the nonhomologous end-joining pathway (NHEJ) is important not only for repair of spontaneous breaks but also for breaks induced in developing lymphocytes during V(D)J (variable [V], diversity [D], and joining [J] genes) recombination of their antigen receptor loci to create a diverse repertoire. Mutations in the NHEJ factor XLF result in extreme sensitivity for ionizing radiation, microcephaly, and growth retardation comparable to mutations in LIG4 and XRCC4, which together form the NHEJ ligation complex. However, the effect on the immune system is variable (mild to severe immunodeficiency) and less prominent than that seen in deficiencies of NHEJ factors ARTEMIS and DNA-dependent protein kinase catalytic subunit, with defects in the hairpin opening step, which is crucial and unique for V(D)J recombination. Therefore, we aimed to study the role of XLF during V(D)J recombination. We obtained clinical data from 9 XLF-deficient patients and performed immune phenotyping and antigen receptor repertoire analysis of immunoglobulin (Ig) and T-cell receptor (TR) rearrangements, using next-generation sequencing in 6 patients. The results were compared with XRCC4 and LIG4 deficiency. Both Ig and TR rearrangements showed a significant decrease in the number of nontemplated (N) nucleotides inserted by terminal deoxynucleotidyl transferase, which resulted in a decrease of 2 to 3 amino acids in the CDR3. Such a reduction in the number of N-nucleotides has a great effect on the junctional diversity, and thereby on the total diversity of the Ig and TR repertoire. This shows that XLF has an important role during V(D)J recombination in creating diversity of the repertoire by stimulating N-nucleotide insertion.
Introduction
DNA double-strand breaks (DSBs) are toxic lesions and, when not corrected, severely affect the differentiation, growth, and function of a cell. DSBs can arise in cells exposed to agents such as ionizing radiation (IR), but are also purposely introduced during development and maturation of T and B lymphocytes. Lymphocytes express a unique antigen receptor on their membrane; namely, the T-cell receptor or immunoglobulin (TR or Ig). Before expression, the loci encoding for these receptors are rearranged through recombination of the multiple variable (V), diversity (D) and joining (J) genes. This process is called V(D)J recombination and ensures the generation of a large diversity of both TR and Ig repertoires.
V(D)J recombination is initiated by recombination activating gene 1 (RAG1) and RAG2 proteins, which introduce DNA DSB near the V, D, and J genes.1,2 These RAG1 and RAG2 proteins are lymphoid specific, but the subsequent repair of the DNA DSBs occurs via the ubiquitously expressed nonhomologous end-joining (NHEJ) DNA repair pathway. After RAG cleavage, the DSBs are recognized by the DNA-dependent protein kinase (DNA-PK) complex, which consists of the DNA-PK catalytic subunit (DNA-PKcs) and the KU70/KU80 heterodimer.3,4 Subsequently, the ARTEMIS protein opens the hairpin-sealed coding ends, which are formed by the RAG proteins.5 Finally, the coding ends are ligated by the DNA ligase IV (LIG4)/XRCC4 complex in conjunction with Cernunnos/XLF.6-9 Recently, a new NHEJ factor called paralog of XRCC4 and XLF has been described. This factor directly interacts with Ku and promotes DNA ligation; however, the exact role of this protein in V(D)J recombination is not yet known.10,11
The diversity of the antigen receptors created during V(D)J recombination has been estimated to be more than 1012. Part of the diversity arises from different combinations of the V, D, and J genes (2 × 106), but to obtain the needed variability in the receptors, the DNA ends are processed during V(D)J recombination. In case of asymmetric opening of the hairpins, palindromic (P) nucleotides can be formed. In addition, nucleotides can be deleted by exonuclease activity, and nontemplated (N) nucleotides can be inserted by terminal deoxynucleotidyl transferase (TdT) at the coupling sites (junctions) of the V, D, and J genes.12 These processing events lead to the junctional diversity of Ig and TR rearrangements, which has an enormous effect on the final repertoire diversity.
Defects in V(D)J recombination block the development of T and B cells, which results in a (severe) combined immunodeficiency.13-15 In addition, patients with defects in NHEJ have increased sensitivity to IR and can present with growth retardation and neurological abnormalities.7,16,17
XLF deficiency in human results in extreme sensitivity to IR, microcephaly, and growth retardation, but the effect on the immune system is variable (mild to severe immunodeficiency).6,7,18 Studies in XLF-deficient mouse models have confirmed the severe sensitivity to IR and chromosomal instability, but the V(D)J recombination defect seemed relatively mild.19,20 So far, the most important action of XLF seems to be the stimulation of LIG4 activity,21,22 probably by stabilizing the XRCC4–LIG4 complex on DNA and promotion of DNA strand alignment.23 However, recently, Lescale et al have shown that the RAG postcleavage complex (RAG-PCC) and XLF have overlapping functions and have suggested that both the RAG-PCC and XLF ensure stabilization of DNA ends after DNA cleavage by RAG.24 To further understand the function of XLF during V(D)J recombination and NHEJ, the characteristics of the immunoglobulin heavy chain (IgH), Igκ (IgK), Igλ (IgL), TR β (TRB), and TR δ (TRD) gene rearrangements were studied in 6 XLF-deficient patients, using next-generation sequencing. We identified an additional role for XLF in the contribution to junctional diversity during V(D)J recombination.
Methods
Cell samples and flow cytometric immunophenotyping
Peripheral blood, bone marrow, and a skin biopsy were obtained with informed consent and according to the guidelines of the local Medical Ethics Committees. Flow cytometric analysis of peripheral blood was performed as previously described.25
Clonogenic survival assay
Clonogenic survival assays with primary skin fibroblasts from patient XLF1 and patient XLF5 were performed as described previously.26 In short, primary skin fibroblasts in exponential growth were trypsinized, and 500 to 2000 cells (5000-80 000 cells for the highest doses) were seeded into 10-cm plastic dishes (2 dishes per dose) and irradiated at room temperature at a dose of approximately 1 Gy/min. After 12 to 14 days, the cells were rinsed with 0.9% NaCl and stained with 0.25% methylene blue for survival assessment. Two independent survival experiments were performed. The results were compared with fibroblasts obtained from patients with defects in ARTEMIS (n = 7), LIG4 (n = 1), and XRCC4 (n = 1).27,28
Analysis of DH–JH junctions from bone marrow and IgH, IgK, IgL, and TRD junctions from peripheral blood
DNA was isolated from bone marrow mononuclear cells or peripheral blood mononuclear cells. DH–JH coding joints were amplified by PCR with family-specific DH primers and a consensus JH primer, as previously described,29 followed by cloning of the PCR products into pGEM-T Easy (Promega). The IgH, IgK, IgL, and TRD junctions were amplified from post-Ficoll peripheral blood (PB) mononuclear cells in a multiplex PCR, using the BIOMED-2 VH1-6 FR1 forward and JH consensus reverse primer (IgH), Vk2 and Vk3 forward and JK1-5 reverse primers (IgK), Vl1/2 and Vl3 forward and Jl1/2/3 reverse primers (IgL), and Vd2 forward and Jd1 reverse primers (TRD).30 Primes were adapted for 454 sequencing by adding the forward A or reverse B adaptor, the “TCAG” key, and multiplex identifier (MID) adaptor. For Illumina sequencing, the primers were adapted by adding a MID adaptor to the forward or the reverse primer. PCR products were purified by gel extraction (Qiagen, Valencia, CA) and Agencourt AMPure XP beads (Beckman Coulter, Fullerton, CA). Subsequently, the PCR concentration was measured using the Quant-it PicoGreen dsDNA assay (Invitrogen, Carlsbad, CA). The purified PCR products were sequenced either on the 454 GS junior instrument (IgH and part of IgK) according the manufacturer’s recommendations, using the GS junior Titanium emPCR kit (Lib-A), sequencing kit, and PicoTiterPlate kit (454 Life Sciences, Roche, Brandford, CT), or using the MiSeq sequencer (IgL, TRD, and part of IgK), using low-complexity 300-base pair paired-end sequencing (V3 chemistry).
The reads were uploaded to IMGT HighV-Quest.31 Subsequently, these output files were analyzed using the IGGalaxy tool and R studio.32,33 For the analysis, we only used the unique productive or unique unproductive rearrangements, in which unique is defined as junctions having the same V, same J, and same nucleotide sequence of the CDR3. Information about the composition of the junctional regions was extracted from the data provided by IMGT HighV-Quest. The majority of the TRD rearrangements we analyzed contained only 1 D gene. Therefore, we used only the VDJ rearrangements, except for the analysis used for Figure 4, in which it is indicated that we analyzed either VDJ or VDDJ rearrangements.
TRB analysis
Junction analysis in Xlf-deficient mouse lymphocytes
TdT expression
Statistical analysis
Differences in numbers of N- and P-nucleotides and deletions were analyzed using the nonparametric Mann-Whitney U test (2-tailed; P < .05 was considered significant) in the GraphPad Prism program (GraphPad Software).
Results
XLF deficiency results in hypersensitivity for ionizing radiation, but relative moderate immunodeficiency
So far, 15 XLF-deficient patients have been described in the literature.6,7,18,37-39 In this study, we describe 9 additional patients (Table 1). All patients (except P3)18 had microcephaly and growth retardation. The XLF-deficient patients suffered from recurrent (opportunistic) infections, but in contrast to patients with “classical” severe combined immunodeficiency with defects in RAG or ARTEMIS, in whom the outcome is usually fatal within the first year of life, unless they receive hematopoietic stem cell transplantation (SCT), the XLF-deficient patients survived beyond the first year of life without SCT. In fact, the 12 patients described in literature and in this study in which immunophenotyping was performed were between 11 months and 14 years old at evaluation (Table 2). However, the numbers of lymphocytes, as well as CD4+ and CD8+ T cells, were reduced or low. The B cells were strongly reduced or absent, especially in the older patients (Table 2).
Clinical characteristics of XLF-deficient patients
| Patient . | Mutation allele 1 . | Allele 2 . | Microcephaly . | Growth retardation . | Infections . | Autoimmunity . | Outcome . |
|---|---|---|---|---|---|---|---|
| P17 | p.R57G | p.C123R | + | + | Bacterial and opportunistic | + | Died at 18 y: septic shock |
| P27 | p.R178X | p.R178X | + | + | Bacterial and opportunistic | − | Died at 4 y: septic shock |
| P37 | c.177delC, c.177+3A>T | c.177delC, c.177+3A>T | + | + | Bacterial and opportunistic | + | Alive: 14 y |
| P47 | c.177delC, c.177+3A>T | c.177delC, c.177+3A>T | + | + | Bacterial and opportunistic | − | Alive: 3 y |
| P57 | p.R57G | p.R57G | + | + | Recurrent respiratory tract | − | Alive: 9 y old |
| P638 | p.Y167X | p.Y167X | + | ND | ND | ND | ND |
| P738 | p.R57X | p.R57X | + | ND | ND | ND | ND |
| P838,39 | g.del ex2-5 | g.del ex2-5 | + | + | ND | ND | ND |
| 2BN6 | p.E5GfsX44 | p.E5GfsX44 | ND | ND | ND | ND | Received SCT |
| P118 | p.R57G | p.R57G | + | + | Recurrent respiratory tract | − | Alive: 12 y |
| P218 | p.D166RfsX2 | 1.9kb deletion | + | + | Urinary tract | − | Alive: 6 y |
| P318 | p.D166RfsX2 | 1.9kb deletion | − | + | Recurrent respiratory tract | − | Alive: 2 y |
| P418 | p.R176X | 6.9kb deletion | + | + | − | Alive: 8 y | |
| P518 | p.R178X | p.R178X | + | + | Mouth lesions | + | Died at 1.5 y: septic shock |
| Patient37 | p.R178X | p.R178X | + | + | Recurrent perianal abscess | Alive: 3 y: SCT planned | |
| XLF1 | p.Y167X | p.Y167X | + | + | Recurrent respiratory tract | − | Alive: SCT at 8 y |
| XLF2 | p.R178X | p.R178X | + | + | Diarrhea | − | Alive: SCT at 1.5 mo |
| XLF3 | p.R57X | p.R57X | + | + | Urinary and respiratory tract | + | Alive: SCT at 15 mo |
| XLF4 | p.R178X | p.R178X | + | + | Respiratory tract | − | Alive SCT at 10 y |
| XLF5 | p.R109AfsX3 | p.R109AfsX3 | + | + | Alive 8 y: SCT planned | ||
| XLF6-1 | p.R178X | p.R178X | + | + | Recurrent respiratory fungal | − | Alive 2 y: SCT planned |
| XLF6-2 | p.R178X | p.R178X | + | + | Recurrent respiratory tract | − | Alive 8 y: SCT planned |
| XLFP1 | p.R178X | p.R178X | + | + | Diarrhea, respiratory tract | − | Alive: SCT at 9 y |
| XLFP2 | p.R178X | p.R178X | + | + | Opportunistic (BCGitis) and bacterial (otitis) | + | Alive: SCT at 12 mo |
| Patient . | Mutation allele 1 . | Allele 2 . | Microcephaly . | Growth retardation . | Infections . | Autoimmunity . | Outcome . |
|---|---|---|---|---|---|---|---|
| P17 | p.R57G | p.C123R | + | + | Bacterial and opportunistic | + | Died at 18 y: septic shock |
| P27 | p.R178X | p.R178X | + | + | Bacterial and opportunistic | − | Died at 4 y: septic shock |
| P37 | c.177delC, c.177+3A>T | c.177delC, c.177+3A>T | + | + | Bacterial and opportunistic | + | Alive: 14 y |
| P47 | c.177delC, c.177+3A>T | c.177delC, c.177+3A>T | + | + | Bacterial and opportunistic | − | Alive: 3 y |
| P57 | p.R57G | p.R57G | + | + | Recurrent respiratory tract | − | Alive: 9 y old |
| P638 | p.Y167X | p.Y167X | + | ND | ND | ND | ND |
| P738 | p.R57X | p.R57X | + | ND | ND | ND | ND |
| P838,39 | g.del ex2-5 | g.del ex2-5 | + | + | ND | ND | ND |
| 2BN6 | p.E5GfsX44 | p.E5GfsX44 | ND | ND | ND | ND | Received SCT |
| P118 | p.R57G | p.R57G | + | + | Recurrent respiratory tract | − | Alive: 12 y |
| P218 | p.D166RfsX2 | 1.9kb deletion | + | + | Urinary tract | − | Alive: 6 y |
| P318 | p.D166RfsX2 | 1.9kb deletion | − | + | Recurrent respiratory tract | − | Alive: 2 y |
| P418 | p.R176X | 6.9kb deletion | + | + | − | Alive: 8 y | |
| P518 | p.R178X | p.R178X | + | + | Mouth lesions | + | Died at 1.5 y: septic shock |
| Patient37 | p.R178X | p.R178X | + | + | Recurrent perianal abscess | Alive: 3 y: SCT planned | |
| XLF1 | p.Y167X | p.Y167X | + | + | Recurrent respiratory tract | − | Alive: SCT at 8 y |
| XLF2 | p.R178X | p.R178X | + | + | Diarrhea | − | Alive: SCT at 1.5 mo |
| XLF3 | p.R57X | p.R57X | + | + | Urinary and respiratory tract | + | Alive: SCT at 15 mo |
| XLF4 | p.R178X | p.R178X | + | + | Respiratory tract | − | Alive SCT at 10 y |
| XLF5 | p.R109AfsX3 | p.R109AfsX3 | + | + | Alive 8 y: SCT planned | ||
| XLF6-1 | p.R178X | p.R178X | + | + | Recurrent respiratory fungal | − | Alive 2 y: SCT planned |
| XLF6-2 | p.R178X | p.R178X | + | + | Recurrent respiratory tract | − | Alive 8 y: SCT planned |
| XLFP1 | p.R178X | p.R178X | + | + | Diarrhea, respiratory tract | − | Alive: SCT at 9 y |
| XLFP2 | p.R178X | p.R178X | + | + | Opportunistic (BCGitis) and bacterial (otitis) | + | Alive: SCT at 12 mo |
ND, not described.
Immunological characteristics of XLF-deficient patients
| Patient . | Lymphocytes . | T cells . | CD4+ T cells . | CD8+ T cells . | B cells . | NK cells . | Age at evaluation . |
|---|---|---|---|---|---|---|---|
| P17 | 0.8(1.4-3.3) | 0.6(1.0-2.2) | 0.4(0.5-1.3) | 0.2(0.3-0.9) | 0(0.1-0.6) | 14 y | |
| P27 | 1.2(2.3-5.4) | 0.7(1.4-3.7) | 0.2(0.7-1.5) | 0.6 (0.4-1.1) | 0.08(0.4-1.4) | 0.3 (0.09-0.9) | 2 y |
| P37 | 1.6 (1.4-3.3) | 0.9(1.0-2.2) | 0.5 (0.5-1.3) | 0.4 (0.3-0.9) | 0(0.1-0.6) | 0.4 (0.04-0.7) | 13 y |
| P47 | 2.6 (2.3-5.4) | 0.6(1.4-3.7) | 0.3(0.7-1.5) | 0.08(0.4-1.1) | 0.2(0.4-1.4) | 1.1 (0.09-0.9) | 2 y |
| P57 | 1.1(1.9-3.7) | 0.7(1.2-2.6) | 0.3(0.5-1.3) | 0.3 (0.3-0.9) | 0.04(0.1-0.6) | 0.2 (0.08-0.7) | 7 y |
| P6 | 0.6(2.3-5.4) | 0.08(0.4-1.4) | 6 y | ||||
| P7 | 1.3(1.9-3.7) | 0.04(0.1-0.6) | 2 y | ||||
| P8 | 0.2(1.4-3.3) | 0(0.1-0.6) | 11 y | ||||
| XLF1 | 1.4(1.7-6.9) | 1.1 (0.9-4.5) | 0.6 (0.5-2.4) | 0.4 (0.3-1.6) | 0.06(0.2-2.1) | 0.2 (0.1-1.0) | 4 y |
| XLF5 | 0.7(1.1-5.9) | 0.5(0.7-4.2) | 0.3 (0.3-2.0) | 0.2(0.3-1.8) | 0(0.2-1.6) | 8 y | |
| XLF6-1 | 1.5(2.6-10.4) | 0.2(1.6-6.7) | 0(1.0-4.6) | 0.1(0.4-2.1) | 0.04(0.6-2.7) | 0(0.2-1.2) | 11 mo |
| XLF6-2 | 0.9(1.1-5.9) | 0.9 (0.7-4.2) | 0.3 (0.3-2.0) | 0.4 (0.3-1.8) | 0.01(0.2-1.6) | 0.1 (0.09-0.9) | 8 y |
| Patient . | Lymphocytes . | T cells . | CD4+ T cells . | CD8+ T cells . | B cells . | NK cells . | Age at evaluation . |
|---|---|---|---|---|---|---|---|
| P17 | 0.8(1.4-3.3) | 0.6(1.0-2.2) | 0.4(0.5-1.3) | 0.2(0.3-0.9) | 0(0.1-0.6) | 14 y | |
| P27 | 1.2(2.3-5.4) | 0.7(1.4-3.7) | 0.2(0.7-1.5) | 0.6 (0.4-1.1) | 0.08(0.4-1.4) | 0.3 (0.09-0.9) | 2 y |
| P37 | 1.6 (1.4-3.3) | 0.9(1.0-2.2) | 0.5 (0.5-1.3) | 0.4 (0.3-0.9) | 0(0.1-0.6) | 0.4 (0.04-0.7) | 13 y |
| P47 | 2.6 (2.3-5.4) | 0.6(1.4-3.7) | 0.3(0.7-1.5) | 0.08(0.4-1.1) | 0.2(0.4-1.4) | 1.1 (0.09-0.9) | 2 y |
| P57 | 1.1(1.9-3.7) | 0.7(1.2-2.6) | 0.3(0.5-1.3) | 0.3 (0.3-0.9) | 0.04(0.1-0.6) | 0.2 (0.08-0.7) | 7 y |
| P6 | 0.6(2.3-5.4) | 0.08(0.4-1.4) | 6 y | ||||
| P7 | 1.3(1.9-3.7) | 0.04(0.1-0.6) | 2 y | ||||
| P8 | 0.2(1.4-3.3) | 0(0.1-0.6) | 11 y | ||||
| XLF1 | 1.4(1.7-6.9) | 1.1 (0.9-4.5) | 0.6 (0.5-2.4) | 0.4 (0.3-1.6) | 0.06(0.2-2.1) | 0.2 (0.1-1.0) | 4 y |
| XLF5 | 0.7(1.1-5.9) | 0.5(0.7-4.2) | 0.3 (0.3-2.0) | 0.2(0.3-1.8) | 0(0.2-1.6) | 8 y | |
| XLF6-1 | 1.5(2.6-10.4) | 0.2(1.6-6.7) | 0(1.0-4.6) | 0.1(0.4-2.1) | 0.04(0.6-2.7) | 0(0.2-1.2) | 11 mo |
| XLF6-2 | 0.9(1.1-5.9) | 0.9 (0.7-4.2) | 0.3 (0.3-2.0) | 0.4 (0.3-1.8) | 0.01(0.2-1.6) | 0.1 (0.09-0.9) | 8 y |
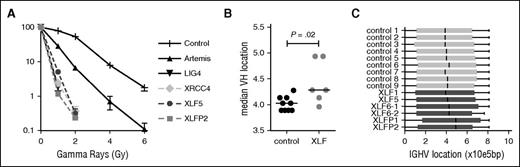
In line with previous studies,6,40 the fibroblasts from patient XLF5 and patient XLFP2 were highly sensitive to IR (Figure 1A). The sensitivity was comparable to LIG4 and XRCC4 deficiency. Interestingly, fibroblasts of ARTEMIS-deficient patients are less IR sensitive.
Clonogenic survival assay and VH gene usage. (A) Clonogenic survival after ionizing radiation showed hypersensitivity of the XLF-deficient patients (XLF5 and XLFP2) compared with control, ARTEMIS-deficient (n = 7), LIG4-deficient (n = 1), and XRCC4-deficient (n = 1) fibroblasts. (B) Median VH gene location according to the relative position in the locus starting from the first functional VH gene (VH4-74), in base pairs (bp). (C) Box-and-whisker representation of VH gene location.
Clonogenic survival assay and VH gene usage. (A) Clonogenic survival after ionizing radiation showed hypersensitivity of the XLF-deficient patients (XLF5 and XLFP2) compared with control, ARTEMIS-deficient (n = 7), LIG4-deficient (n = 1), and XRCC4-deficient (n = 1) fibroblasts. (B) Median VH gene location according to the relative position in the locus starting from the first functional VH gene (VH4-74), in base pairs (bp). (C) Box-and-whisker representation of VH gene location.
These data suggest that XLF is crucial for DNA DSBs in cells, given the strongly increased sensitivity to IR in XLF deficiency and that the absence of XLF results in reduced B and T cells.
Antigen receptor repertoire analysis in XLF-deficient patients
To further investigate the V(D)J recombination defect in XLF-deficient patients, their antigen receptor repertoire analysis was performed. Vera et al have shown previously that both Xlf knockout mice and an XLF-deficient patient had a skewing of the TRA repertoire toward the more 3′ (proximal) V and J genes.20 They hypothesized that the reduced thymocyte lifespan does not allow the T cells to undergo multiple waves of VαJα rearrangements, which can be necessary for positive selection of T cells. At the IgH and TRB loci, these subsequent rearrangements do not occur, so we wondered whether the combinational diversity was also affected in these rearrangements. Therefore, we performed antigen receptor repertoire analysis by next-generation sequencing of the IgH and TRB rearrangements. Overall, the median VH gene location was more downstream (proximal) in the XLF-deficient patients (Figure 1B); however, this seemed mainly caused by skewing of the VH gene location in patients XLFP1 and XLFP2, as the other patients are in the same range as the healthy controls (Figure 1C). TRB rearrangements in XLF5 seem slightly skewed toward 5′ (distal) V genes (supplemental Figure 1A, available on the Blood Web site). The IgH and TRB repertoires of XLF-deficient patients showed a diverse pattern of V, D, and J genes (supplemental Figure 1B; supplemental Figure 2), suggesting that except for TRA, the combinational diversity is intact in XLF deficiency.
Composition of the junctions in an XLF-deficient patient and Xlf-deficient mice is changed
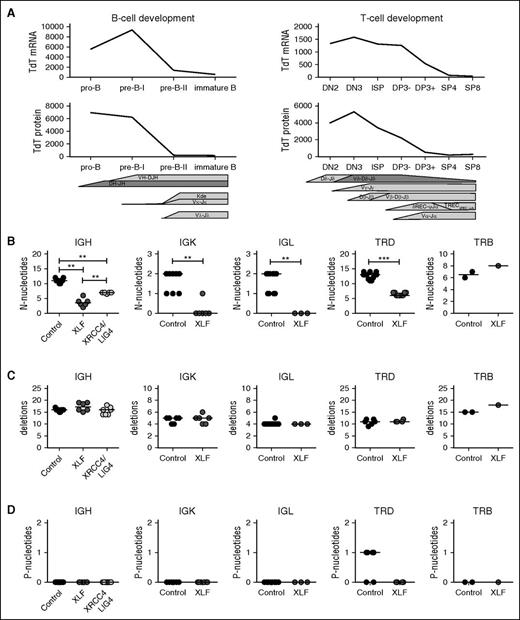
In addition to combinational diversity, the total diversity of the antigen receptors is also heavily influenced by junctional diversity. Therefore, we studied DH–JH junction characteristics in XLF-deficient bone marrow-derived precursor B cells. Recombination of the DH gene to the JH gene is the first rearrangement occurring during IgH recombination, and is therefore suitable to study the V(D)J recombination process in recombination-deficient patients. Interestingly, DH-JH junctions in XLF-deficient patient XLF1 had normal numbers of deletions and P-nucleotides, but strongly reduced numbers (0.1 vs 8.1 in controls) of N-nucleotides (supplemental Figure 3A). This reduction was larger and significantly different from that in a LIG4-deficient patient. In contrast to XLF1, the LIG4 patient had a strong increase in the number of deletions, which explains the lower number of N-nucleotides.
In addition, we reanalyzed junctions derived from Xlf-deficient mouse lymphocytes that were previously published.19,20 Although it was previously suggested that the junctions in Xlf-deficient mice were normal, both the junctions derived from thymocytes and the IgH junctions had a significant decrease of 1.8 (P = .0109) and 1.3 (P = .011) N-nucleotides, respectively (supplemental Figure 3B).
Analysis of TR and Ig rearrangements, using next-generation sequencing
To further assess the role of XLF in junctional diversity, we analyzed V(D)J junctions using next-generation sequencing and evaluated 3 junctional region characteristics: the number of N-nucleotides, which are inserted by TdT; the number of nucleotide deletions; and the number of P-nucleotides, which arise as a result of asymmetric hairpin opening by ARTEMIS. For correct interpretation of the number of N-nucleotides, it is important to note that TdT mRNA and protein expression is highest during early rearrangements (IgH, TRD, TRG) (Figure 2A), and lower during late rearrangements (IgK, IgL, and TRB). All rearrangements, except for TRB, were studied at the DNA level, allowing us to study both the productive rearrangements and the unproductive rearrangements. These unproductive rearrangements are out of frame or contain a stop codon, and consequently have not been selected. Depending on the availability of the patient material, we used 1 to 6 patients and 2 to 10 age-matched controls per rearrangements. We analyzed unique junctions, which we defined as a unique combination of V, J, and nucleotide CDR3 sequence. The numbers of unique rearrangements are listed in supplemental Table 1.
TdT expression levels and junction characteristics of productive Ig and TR rearrangements. (A) Expression of TdT is higher during the early Ig rearrangements and TR rearrangements, measured at RNA level by microarray (gene expression)35,36 and at the protein level by flow cytometry. (B) Median number of N-nucleotides, (C) deletions, and (D) P-nucleotides in productive unique IgH, IgK, IgL, TRD, and TRB rearrangements in healthy controls and XRCC4/LIG4- and XLF-deficient patients. The numbers of productive unique rearrangements used to calculate the median are listed in supplemental Table 1. *P ≤ .05; **P ≤ .01; ***P ≤ .001 by 2-tailed Mann-Whitney U test.
TdT expression levels and junction characteristics of productive Ig and TR rearrangements. (A) Expression of TdT is higher during the early Ig rearrangements and TR rearrangements, measured at RNA level by microarray (gene expression)35,36 and at the protein level by flow cytometry. (B) Median number of N-nucleotides, (C) deletions, and (D) P-nucleotides in productive unique IgH, IgK, IgL, TRD, and TRB rearrangements in healthy controls and XRCC4/LIG4- and XLF-deficient patients. The numbers of productive unique rearrangements used to calculate the median are listed in supplemental Table 1. *P ≤ .05; **P ≤ .01; ***P ≤ .001 by 2-tailed Mann-Whitney U test.
Overall, the numbers of N-nucleotides in early rearrangements was much higher than in late rearrangements. The numbers for IgK and IgL rearrangements are also lower, because these are single-step rearrangements. In line with the DH–JH junctions and the junctions derived from Xlf-deficient mice, the median number of N-nucleotides per junction in XLF-deficient patients was significantly decreased in the productive and unproductive IgH, IgK, IgL and TRD (Figures 2B and 3A). Similar to the TRB junctions in mice (supplemental Figure 3B), no difference in the number of N-nucleotides was observed in TRB junctions in XLF-deficient patients.
Junction characteristics of unproductive Ig and TR rearrangements. (A) Median number of N-nucleotides, (B) deletions, and (C) P-nucleotides in unproductive unique IgH, IgK, IgL, TRD, and TRB rearrangements in healthy controls and XRCC4/LIG4- and XLF-deficient patients. The numbers of productive unique rearrangements used to calculate the median are listed in supplemental Table1. *P ≤ .05; **P ≤ .01; ***P ≤ .001 by 2-tailed Mann-Whitney U test.
Junction characteristics of unproductive Ig and TR rearrangements. (A) Median number of N-nucleotides, (B) deletions, and (C) P-nucleotides in unproductive unique IgH, IgK, IgL, TRD, and TRB rearrangements in healthy controls and XRCC4/LIG4- and XLF-deficient patients. The numbers of productive unique rearrangements used to calculate the median are listed in supplemental Table1. *P ≤ .05; **P ≤ .01; ***P ≤ .001 by 2-tailed Mann-Whitney U test.
Because XLF works together with XRCC4 and LIG4 in the ligation complex, we wanted to know whether the reduction of N-nucleotides is specific for XLF deficiency or whether it is related to a defect in the ligation complex. Therefore, we analyzed unique productive and unproductive IgH rearrangements derived from 7 XRCC4- and LIG4-deficient patients. These patients also had a significant reduction in the number of N-nucleotides (7 vs 11 nt in the controls; Figure 2B); however, the reduction was significantly less than observed in XLF-deficient patients (3.5 nt). This suggests that defects in the ligation complex lead to reduced addition of N-nucleotides, but the largest reduction in the number of N-nucleotides is specific for XLF deficiency.
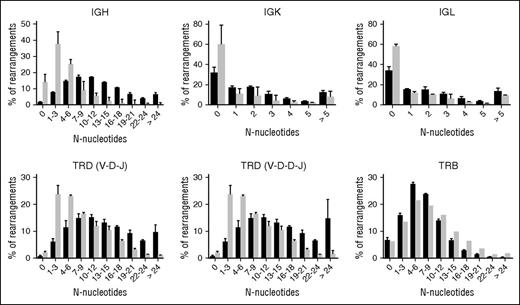
In addition to the reduced number of N-nucleotides, the percentage of rearrangements with a low number of N-nucleotides was increased (Figure 4). In TRD and IgH rearrangements, the percentage of rearrangements that have low numbers of N-nucleotides (1-3 N-nucleotides) was much higher compared with the controls. Also, in the IgK and IgL rearrangements, the percentages of rearrangements with 0 N-nucleotides were much higher (63.4% vs 32.6% and 57.7% vs 33.8%) in XLF-deficient patients (Figure 4). Only in TRB rearrangements was no difference observed. These data imply that the junction diversity of TRγδ and Ig is much lower in XLF-deficient patients.
Number of N-nucleotides per junction. The percentage of productive unique rearrangements with a certain number of total N-nucleotides per junction. The TRD rearrangements were separated on the basis of the presence of 1 D gene (VDJ) or 2 D genes (VDDJ). Controls are indicates in black, and the XLF-deficient patients are indicated in gray.
Number of N-nucleotides per junction. The percentage of productive unique rearrangements with a certain number of total N-nucleotides per junction. The TRD rearrangements were separated on the basis of the presence of 1 D gene (VDJ) or 2 D genes (VDDJ). Controls are indicates in black, and the XLF-deficient patients are indicated in gray.
Reduced N-nucleotides result in a shorter CDR3 length
The diversity and the length of the CDR3 region are influenced by both the insertion of N-nucleotides and the deletion of nucleotides during V(D)J recombination. In contrast to the low numbers of N-nucleotides, the number of deletions and P-nucleotides were normal in all productive and unproductive IgH, IgK, IgL, TRD, and TRB rearrangements (Figures 2C and 3B).
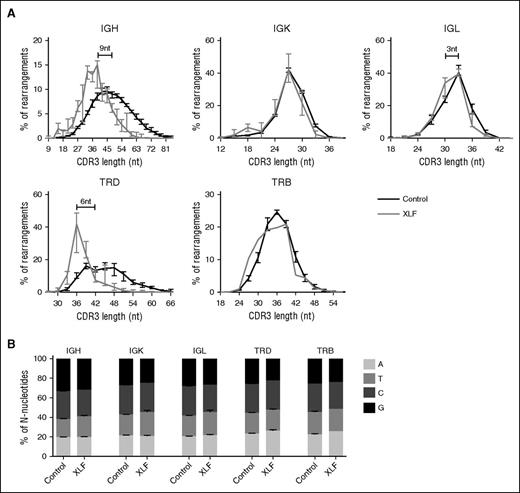
The median CDR3 length in IgH rearrangements is 48 nt, and the median number of N-nucleotides is 11, so the N-nucleotides contribute for approximately a fourth of the total IgH CDR3 length. This holds also true for TRD, where the total number of N-nucleotides can be even higher if multiple D genes are used. The median number of N-nucleotides in TRB rearrangements (6.5 nt) is lower, and it is very low in IgK (2 nt) and IgL (2 nt); therefore, the number of N-nucleotides contributes less to the CDR3 length in these rearrangements. In line with this, we found that the median CDR3 length in IgK and TRB was not reduced and was only 3 nt shorter in IgL rearrangements (Figure 5A). However, the XLF-deficient patients had the strongest reduction in the number of N-nucleotides in IgH and TRD, and as a consequence, the CDR3 length was strongly decreased with approximately 9 nt in IgH and 6 nt in TRD (Figure 5A). This implies that the CDR3 protein sequence is 2 to 3 amino acids shorter. Such reduction in the number of N-nucleotides has a great effect on the junctional diversity, and thereby on the total diversity of the Ig and TRD repertoire.
Distribution of CDR3 length and nucleotide incorporation. (A) The average percentage of productive unique IgH, IgK, IgL, TRD, and TRB rearrangements with a certain nucleotide CDR3 length in XLF-deficient patients (gray) and control (black). The numbers in the IgH and TRD graph indicate the difference in the median CDR3 length between the XLF-deficient patients and the controls. (B) Distribution of the total number of N-nucleotides that are an A, T, C, or G nucleotide in controls and XLF-deficient patients.
Distribution of CDR3 length and nucleotide incorporation. (A) The average percentage of productive unique IgH, IgK, IgL, TRD, and TRB rearrangements with a certain nucleotide CDR3 length in XLF-deficient patients (gray) and control (black). The numbers in the IgH and TRD graph indicate the difference in the median CDR3 length between the XLF-deficient patients and the controls. (B) Distribution of the total number of N-nucleotides that are an A, T, C, or G nucleotide in controls and XLF-deficient patients.
No difference in bias of nucleotide incorporation by TdT
Although TdT adds random nucleotides to single-stranded DNA, the nucleotide incorporation is biased. Several studies have shown that TdT preferentially uses dGTP and dCTP.41,42 Therefore, we analyzed the total number of N-nucleotides per rearrangement and determined the percentage of A, T, C, and G nucleotides. In line with the previous studies, we observed in all rearrangements that more than half of the N-nucleotides were either dGTP or dCTP (Figure 5B). This was not different in the XLF-deficient patients, implying that only the number of N-nucleotides is reduced.
Discussion
XLF is important for general DSB repair and for repair of DSBs during V(D)J recombination; however, the effect of XLF deficiency on both processes seems different. XLF deficiency has a severe effect on DSB repair, as XLF-deficient patients are hypersensitive to IR (Figure 1A) and have a significant 50% reduction of cell survival.7,20 The IR sensitivity in XLF deficiency is similar to XRCC4 and LIG4 deficiency, which belong to the NHEJ ligation complex. Patients deficient for ARTEMIS, which is specifically involved in repair of certain DSBs and in hairpin opening during V(D)J recombination, are less sensitive for IR than XLF-deficient patients (Figure 1A). In line with this, defects in the NHEJ ligation complex give a severe neurological phenotype and growth failure, which is not seen in ARTEMIS and in a DNA-PKcs-deficient patient with a hairpin opening defect.15,29 On the contrary, hairpin opening defects have a severe effect on V(D)J recombination, and consequently give rise to severe combined immunodeficiency, whereas defects in the NHEJ ligation complex give a milder immunodeficiency, which is even completely absent in XRCC4-deficient patients. Most of the XLF-deficient patients survived the first years of life, or even up to 18 years, without SCT. Thus, mutations in the NHEJ ligation complex result in severe neurological complications and growth failure, but a milder immunodeficiency compared with defects in NHEJ factors involved in hairpin opening. The difference in effect on DNA repair and V(D)J recombination in XLF deficiency might be explained by the 2-tier synapse model that has recently been proposed by Lescale et al, showing that both the RAG-PCC and XLF ensure stabilization of DNA ends after RAG1/2 cleavage.24 They showed that the RAG-PCC has functional redundancy with XLF in RAG DSB stabilization, which indicates that in the presence of RAG, DNA end repair can be achieved during V(D)J recombination.
In a previous study, we analyzed the precursor B-cell differentiation in bone morrow of patient XLF143 and showed that XLF1 had considerable numbers of pre-B-II and immature B cells, although the frequency of pro-B and pre-B-I cells was slightly elevated. Thus, in XLF deficiency, the B-cell differentiation is not completely blocked at the pre-B-I stage, where V(D)J recombination takes place, as is the case in RAG, ARTEMIS, and DNA-PKcs deficiency. This was comparable to the fairly normal precursor B-cell compartment in Xlf-deficient mice.19 However, XLF-deficient patients had a reduced number of T and B cells; especially in the older XLF-deficient patients, the numbers of B cells were absent. Studies in mice have shown that hematopoietic stem cell dysfunction may contribute to progressive lymphocytopenia in Xlf−/− mice,44 and that Xlf−/− thymocytes have increased expression of P53-associated proapoptotic genes, which likely makes them more prone to apoptosis.20
In this study, we have shown that XLF deficiency results in a strong reduction in the number of N-nucleotides. The largest reduction of N-nucleotides was observed in IgH and TRD rearrangements, which are early rearrangements taking place during differentiation stages with high TdT expression and where normally many N-nucleotides are added (Figure 2A). This reduction was not caused by increased removal of the N-nucleotides, as the number of deletions was normal. In XRCC4- and LIG4-deficient patients, the number of N-nucleotides was also decreased in IgH rearrangements, but still the total number was significantly higher than in the XLF-deficient patients. These data suggest that the ligation complex is important for N-nucleotide addition, and that XLF plays a dominant role.
During IgK, IgL and TRB (late) rearrangements, TdT expression is lower, and fewer N-nucleotides are added per junction. However, we could still observe a reduction in the number of N-nucleotides in IgK and IgL rearrangements in the XLF-deficient patients. Surprisingly, no reduction in the number of N-nucleotides was observed in TRB rearrangements in the XLF-deficient patients (Figure 2B) and the Xlf-deficient mice (supplemental Figure 3B). In TRB rearrangements, the number of N-nucleotides is lower than in IgH and TRD rearrangements, probably because of lower TdT expression. Similar to IgH rearrangements, TRB rearrangements contain 2 junctional regions in which N-nucleotides are added. The controls had on average 2 to 3 N-nucleotides per junction, whereas TRB rearrangements of patient XLF5 even had a slightly higher number of nucleotides (4 nt) per junction. N-nucleotide addition might be less dependent on XLF during TRB rearrangements. Alternatively, thymocytes undergo stringent selection in the thymus, and therefore only thymocytes expressing TRs with a correct CDR3 length might be selected during positive selection. Finally, we cannot exclude that other proteins such as polλ and polμ play a (different) role during the different Ig and TR rearrangements. Bertocci et al have shown that polλ is involved in heavy chain rearrangements, and polμ in light chain rearrangements.45,46
Recently, Rechavi et al have shown that fetal-derived lymphocytes also have reduced numbers of numbers of N-nucleotides in IgH and TRB rearrangements.47 In addition, they showed preferential use of the DH-proximal VH genes IgHV6-1 and IgHV1-2 and the JH-proximal IgHD7-27 genes in the fetal IgH rearrangements. In contrast, the use of IgHV6-1, IgHV1-2, and IgHD7-27 was normal in the XLF-deficient patients (data not shown), suggesting it is not likely that the XLF-deficient patients have an increased frequency of the fetal-derived lymphocytes. We rather think that XLF is important for the position, recruitment, or activation of TdT; however, the exact mechanism needs to be unraveled. Since its discovery in 2005, several functions have been described for XLF. It stimulates LIG4 activity,21,22 and it can form long filaments with XRCC4 that keep the DNA ends together in a ligation synapse,24,48-51 and XLF is essential for gap-filling by polymerase (pol) λ and polμ during NHEJ. The latter function is interesting, as polλ and polμ belong to the same polX family of polymerases as TdT. Loss of N-nucleotide addition was also observed in TdT−/− and Ku80−/− mice.52-54 Purugganan et al hypothesized that the most likely explanation for the loss of N-nucleotides in the Ku80-deficient mice was loss of TdT recruitment.54 Gel mobility shift assays have also shown that Ku, together with the Xrcc4–Ligase IV complex, is required for the recruitment for the 2 closely related polymerases polμ and polλ,55,56 Ku80 deficiency resulted in complete absence of N-nucleotides, whereas the Xlf-deficient mice only have a reduced number of N-nucleotides.
In this study, we have shown that XLF has a dual role in DSB repair and V(D)J recombination. Although the effect of XLF deficiency seems more moderate in V(D)J recombination, we could show that XLF has an important role during N-nucleotide addition.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The research for this manuscript was in part performed within the framework of the Erasmus Postgraduate School Molecular Medicine. We acknowledge A. P. Jackson, A. W. Langerak, M. Berkowska, and M. Wentink for technical support and advice.
This work was supported by grants from the foundation “Sophia Kinderziekenhuis Fonds” (grant 589) (H.I. and M.v.d.B.), the Netherlands Organization for Scientific Research/the Netherlands Organization for Health Research and Development (grant 91712323) (M.v.d.B.), and the Dutch Cancer Foundation (grant EMCR 2008-4045) (D.C.v.G.).
Authorship
Contribution: H.I. and M.v.d.B. designed the research and wrote the paper; H.I., R.L.W., D.v.Z., R.A.H., I.P.-K., E.S., I.J., R.L., and N.S.V. performed experiments and/or analyzed data; J.R., K.S., A.W., M.L., T.P., and H.H.A. provided patient material, collected clinical data, and critically read the manuscript; and A.P.S., D.C.v.G., and J.J.M.v.D. contributed to essential discussion and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mirjam van der Burg, Erasmus MC, Department of Immunology, Wytemaweg 80, 3015 CN Rotterdam, The Netherlands; e-mail: m.vanderburg@erasmusmc.nl.