Key Points
c-Myb is essential for neutrophil terminal differentiation by targeting granule gene expression.
c-Myb and Cebp1 act cooperatively to regulate neutrophil maturation in zebrafish.
Abstract
Neutrophils are the key effectors for generating innate immunity in response to pathogenic infection and tissue injury in vertebrates. Dysregulation of neutrophil development and function is known to associate with various human disorders. Yet, the genetic network that orchestrates lineage commitment, differentiation, and maturation of neutrophils remains incompletely defined. Here, we present an in vivo study to delineate the genetic program underlying neutrophil development during zebrafish embryonic myelopoiesis. We show that loss of c-Myb function has no effect on macrophages but severely impairs neutrophil terminal differentiation, resulting in the accumulation of neutrophils with unsegmented nuclei and scant granule. This neutrophilic defect, which resembles the neutrophil-specific granule deficiency (SGD) caused by the mutations in CCAAT/enhancer-binding protein ε (C/EBPε) in humans, is attributed, at least in part, to the downregulation of the granule protein transcription. Likewise, genetic inactivation of Cebp1, the zebrafish functional homolog of mammalian C/EBPε, also leads to a similar SGD-like phenotype in zebrafish. Genetic epistasis and biochemical analysis further reveals that c-Myb and Cebp1 act in parallel and cooperatively to control neutrophil differentiation by directly regulating granule protein gene transcription. Our study indicates that c-MYB is an intrinsic master regulator for neutrophil terminal differentiation and a potential target in SGD patients.
Introduction
Neutrophils are the most abundant white blood cells in the human body and are essential for removing pathogens such as bacteria and fungi to prevent human infection. On the other hand, hyperactivation of neutrophils can also cause chronic inflammatory damage to tissues.1-4 Because the numbers and activities of neutrophils are tightly controlled, dysregulation of neutrophil development and function can lead to the onset and progression of a variety of human disorders including neutropenia, neutrophil-specific granule deficiency (SGD), chronic granulomatous disease, and leukemia.1-4
The development of neutrophils has been best characterized in mice.5 In adult mice, the earliest myeloid-restricted precursors derived from hematopoietic stem/progenitor cells are granulocyte-macrophage progenitors (GMPs). The multipotent granulocyte-macrophage progenitors subsequently differentiate into neutrophil-monocyte progenitors, which further commit to the earliest neutrophilic progenitors termed myeloblasts/promyelocytes. The myeloblasts then differentiate into myelocytes which finally undergo terminal differentiation to give rise to mature neutrophils. Genetic and biochemical studies in mice and cell lines have indicated that neutrophil development is mainly governed by cell type–specific transcription factors including the Ets transcription factor PU.1, Runt domain protein RUNX1, and the CCAAT/enhancer-binding protein α (C/EBPα); C/EBPβ and C/EBPε are the best-known transcription factors involved in the lineage commitment and subsequent differentiation and maturation of neutrophils.1-4
A hallmark of mature neutrophils is the presence of granules in their cytoplasm.6,7 There are 3 types of granules: primary, secondary, and tertiary granules, which share common features in structure, but substantial differences in protein contents. The primary granules are the first granules to appear and they are peroxidase positive. As neutrophil precursors undergo further differentiation, the profile of granule proteins synthesized in the differentiating cells changes, which leads to the formation of peroxidase-negative secondary and tertiary granules. The secondary are identified by a high content of lactoferrin, whereas the tertiary are enriched in gelatinase. Although the biological significance of the formation of various granule subtypes remains incompletely understood, they appear to display different accessibility for exocytosis8 and inflammatory responses.9-11 The heterogeneity of neutrophil granules can be partially explained by targeting-by-timing hypothesis, meaning that targeting of protein into a certain subset of granules is determined by the time window of their synthesis.12,13 Yet, the molecular basis controlling granule protein expression remains poorly defined. To date, C/EBPε is the best characterized transcription factor known to be involved in the granule protein expression. Targeted deletion of C/EBPε in mice results in the production of defective neutrophils with immature nuclei and absent or low levels of several secondary granule protein.14,15 Likewise, mutations of C/EBPε in humans cause SGD, in which neutrophils display bilobed nuclei and lack secondary and tertiary granules.16 However, some SGD patients contain intact C/EBPε alleles and express normal levels of C/EBPε proteins,17 indicating that SGD in these patients is caused by other unknown genetic lesions.
c-MYB is the cellular homolog of v-MYB oncogene involved in acute lymphocytic leukemia and myeloid leukemia.18 The physiological role of c-MYB is unveiled by scrutinizing a cohort of c-MYB mutant alleles. Phenotype analysis of c-Myb–null mice indicates that c-MYB is essential for definitive hematopoiesis.19 Lineage-specific roles for c-MYB are uncovered by several c-MYB conditional and hypomorphic alleles which suggest that c-MYB positively regulates erythroid and lymphoid development but negatively regulates megakaryocyte and adult hematopoietic stem cells (HSCs).20-25 Interestingly, except for lack of eosinophils in c-MYBM303V mice,24 myeloid lineage defects appear to be subtle in these viable c-MYB mutant alleles, perhaps due to inadequate suppression of c-MYB activity and lack of in-depth analysis in these alleles.
Integrating the advantages for both embryological and genetic analysis, zebrafish have emerged as a prominent model for study of vertebrate development and human disorders.26 In particular, the zebrafish embryonic myelopoiesis arises directly from the rostral blood island (RBI) and produces macrophages and neutrophils,27-32 allowing for direct assessment of the roles of transcription factors in myeloid development in the absence of their potentially confounding epistatic requirement in the earlier hematopoietic progenitors. Here, we describe a regulatory network showing that during zebrafish embryonic myelopoiesis the neutrophil terminal differentiation is differentially controlled by c-Myb and Cebp1. We further demonstrate that c-Myb and Cebp1 act in parallel and cooperatively to govern neutrophil terminal maturation by impinging on the transcriptional expression of granule proteins.
Methods
Zebrafish husbandry
Histology
Zebrafish embryo dissociation, FACS, and May-Grünwald-Giemsa staining
Embryo dissociation and fluorescence-activated cell sorter (FACS) were performed as described previously.38-40 Because Tg(coro1a:GFP) specifically labels the myeloid population in zebrafish,35 Tg(coro1a:GFP)+/+;c-mybhkz3/+ were outcrossed with Tg(lyz:Dsred)+/+;c-mybhkz3/+ and the resulting c-mybhkz3/hkz3 mutants and siblings were separated based on green fluorescent protein (GFP) and Dsred: GFP+ single-positive embryos were c-mybhkz3/hkz3 mutants, whereas GFP+Dsred+ double-positive embryos represented siblings. The cebp1smu1/smu1 embryos were collected by intercrossing Tg(coro1a:GFP)+/+;cebp1smu1/smu1, whereas siblings were collected by intercrossing the same clutch of Tg(coro1a:GFP)+/+. The GFP+ cells of each group were collected from totally ∼5000 embryos using MoFlo XDP (Beckmann) (∼1000 embryos once, performed 5 times). Neutrophils were distinguished from macrophages by morphology by May-Grünwald-Giemsa staining with cytospin-collected GFP+ cells on slides.41
Transient GFP reporter assay
pTol-lyz:GFP or pTol-lyz:GFP with mutated c-Myb or/and Cebp1 recognition sites were injected into 1-cell stage AB embryos at the dose of 100 pg per embryo. Activity of the GFP reporter was scored by counting the number of GFP+ cells at 2 days postfertilization (dpf).
Luciferase reporter assays
lyz luciferase reporter (150 ng), c-myb or/and cebp1 expression (750 ng), and Renilla constructs (5 ng) were cotransfected into 293T cells (per well in 12-well plate) and luciferase activity was measured according to the Dual Luciferase Assay kit (Promega) instructions.
Chromatin immunoprecipitation
Capped 6×Myc-c-myb messenger RNA (mRNA) was injected into 1-cell stage embryos (100 pg per embryo). Six hundred injected embryos were harvested at 8 hours postfertilization (hpf) for brief fixation. Crosslinked chromatin was immunoprecipitated with anti-Myc antibody or anti-Dsred antibody (negative control) according to the procedure described by Hart et al.42 The immunoprecipitates were subjected to semiquantitative polymerase chain reaction (PCR).
Electrophoretic mobility shift assay
For gel-shift assay, 200 ng of poly(dI-dC), 2 pmol FAM-labeled probes and 3 μg of purified protein were mixed in binding buffer43 in a total 20-μL reaction and incubated at 4°C for 2 hours. The mixture was then loaded onto a 6% polyacrylamide gel (0.5× Tris/Borate/EDTA) for electrophoresis (160 V at 4°C). For the competition studies, 250-fold molar excess of unlabeled competitor oligonucleotides was added to the binding reactions. Gels were screened by Typhoon Trio scanner (GE Healthcare).
Microscopy and imaging
Video-enhanced (VE) differential interference contrast (DIC) microscopy was done with a 60×/1.00 numerical aperture water-immersion objective mounted on an Olympus BX51 microscope as previously described.27 VE-DIC images were captured with a DP71 Olympus color camera and recorded. Whole-mount or magnified bright-field images were taken using a Zeiss AxioCam HRc camera mounted on a Zeiss Discovery V20 microscope. Fluorescent images were captured with Olympus FV1000 confocal microscopy.
Statistical analysis
Data were analyzed by SPSS software (version 20) using the Student t test for comparisons between 2 groups and 1-way analysis of variance (ANOVA; with Bonferonni or Dunnett T3 posttest adjustment) among multiple groups. Significance was accepted when P < .05. Data are expressed as mean ± standard error of the mean (SEM).
Results
Suppression of c-Myb function in zebrafish blocks neutrophil maturation
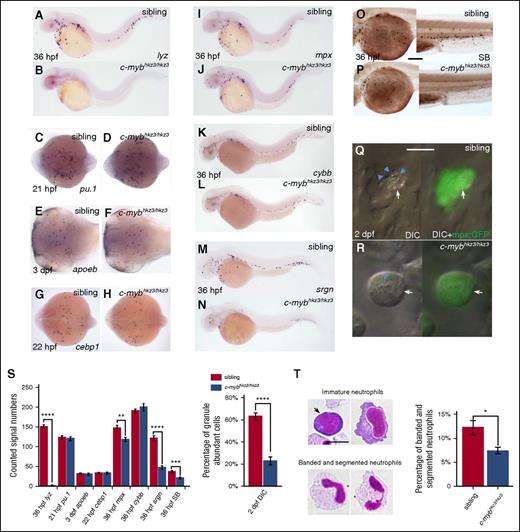
To identify new molecular determinants involved in neutrophil development, we turned our attention to c-mybhkz3/hkz3zebrafish mutants, which were originally isolated as hematopoiesis-defective mutants lacking lysozyme c (lyz), a key component of granule proteins predominantly expressed in neutrophils.37,44-46 This mutant harbors a splicing mutation that results in the synthesis of a truncated c-Myb protein which lacks the transactivation domain but retains the DNA-binding domain.37 Our previous study has shown that, similar to the c-MYB knockout mice,19 the c-mybhkz3/hkz3 mutants fail to develop fetal and adult hematopoiesis due to the impairment of HSCs.37 However, the absence of lyz expression at early developmental stages in c-mybhkz3/hkz3 mutants (Figure 1A-B) suggested that inactivation of c-Myb might cause a severe defect in embryonic myelopoiesis because the majority of myeloid cells that emerged before 3 dpf are derived from embryonic myelopoietic tissue, the RBI.32 We therefore examined embryonic myelopoiesis in c-mybhkz3/hkz3 mutants with myeloid progenitor and macrophage linage markers and found that the formation of myeloid progenitors and their progression into the macrophage branch were not affected by the c-mybhkz3/hkz3 mutation as evidenced by normal pu.1 expression in myeloid progenitors at 21 hpf (Figure 1C-D) and apoeb expression in microglia, the brain-resident macrophages at 3 dpf (Figure 1E-F). These observations suggest that embryonic neutrophils, but not macrophages, are severely impaired in c-mybhkz3/hkz3 mutants.
Neutrophil differentiation is severely impaired in c-mybhkz3/hkz3 mutants. (A-B) Loss of lyz expression in c-mybhkz3/hkz3 mutants. WISH of lyz in 36-hpf siblings (A) and c-mybhkz3/hkz3 mutants (B). (C-H) Myeloid progenitor, macrophage lineage, and neutrophil progenitor markers were not affected in c-mybhkz3/hkz3 mutants as evidenced by unaltered expression of pu.1 (C-D), apoeb (E-F), and cebp1 (G-H). (I-J) mpx+ cells were slightly decreased in c-mybhkz3/hkz3 mutants. WISH of mpx expression in 36-hpf siblings (I) and c-mybhkz3/hkz3 mutants (J). (K-L) Unaltered cybb expression in c-mybhkz3/hkz3 mutants. WISH of cybb expression in 36-hpf siblings (K) and c-mybhkz3/hkz3 mutants (L). (M-N) Decreased srgn expression in c-mybhkz3/hkz3 mutants. WISH of srgn expression in 36-hpf siblings (M) and c-mybhkz3/hkz3 mutants (N). (O-P) Decreased SB+ cells in c-mybhkz3/hkz3 mutants. SB staining in 36-hpf siblings (O) and c-mybhkz3/hkz3 mutants (P). (Q-R) In vivo imaging of 2-dpf Tg(mpx:GFP) embryos by VE DIC microscopy. Left panel, Bright-field DIC image; right panel, an overlay of bright-field DIC and fluorescent image. Blue arrowheads identify granules contained by neutrophils (white arrows) expressing GFP. (S) Quantifications of WISH, SB staining, and DIC analysis for panels A to R (Student t test, **P < .01, ***P < .001, ****P < .0001; WISH and SB, n ≥ 20; DIC, n ≥ 60). (T) May-Grünwald-Giemsa staining of neutrophils in 3-dpf embryos (left) and neutrophils were quantitated by morphology (right) (Student t test, *P < .05, n ≥ 500). Scale bars, 200 μm (O-P) and 5 μm (Q, R, T).
Neutrophil differentiation is severely impaired in c-mybhkz3/hkz3 mutants. (A-B) Loss of lyz expression in c-mybhkz3/hkz3 mutants. WISH of lyz in 36-hpf siblings (A) and c-mybhkz3/hkz3 mutants (B). (C-H) Myeloid progenitor, macrophage lineage, and neutrophil progenitor markers were not affected in c-mybhkz3/hkz3 mutants as evidenced by unaltered expression of pu.1 (C-D), apoeb (E-F), and cebp1 (G-H). (I-J) mpx+ cells were slightly decreased in c-mybhkz3/hkz3 mutants. WISH of mpx expression in 36-hpf siblings (I) and c-mybhkz3/hkz3 mutants (J). (K-L) Unaltered cybb expression in c-mybhkz3/hkz3 mutants. WISH of cybb expression in 36-hpf siblings (K) and c-mybhkz3/hkz3 mutants (L). (M-N) Decreased srgn expression in c-mybhkz3/hkz3 mutants. WISH of srgn expression in 36-hpf siblings (M) and c-mybhkz3/hkz3 mutants (N). (O-P) Decreased SB+ cells in c-mybhkz3/hkz3 mutants. SB staining in 36-hpf siblings (O) and c-mybhkz3/hkz3 mutants (P). (Q-R) In vivo imaging of 2-dpf Tg(mpx:GFP) embryos by VE DIC microscopy. Left panel, Bright-field DIC image; right panel, an overlay of bright-field DIC and fluorescent image. Blue arrowheads identify granules contained by neutrophils (white arrows) expressing GFP. (S) Quantifications of WISH, SB staining, and DIC analysis for panels A to R (Student t test, **P < .01, ***P < .001, ****P < .0001; WISH and SB, n ≥ 20; DIC, n ≥ 60). (T) May-Grünwald-Giemsa staining of neutrophils in 3-dpf embryos (left) and neutrophils were quantitated by morphology (right) (Student t test, *P < .05, n ≥ 500). Scale bars, 200 μm (O-P) and 5 μm (Q, R, T).
Previous study has defined embryonic neutrophil development into 3 successive stages: the cebp1+ early neutrophilic progenitors, the mpx+/lyz+ intermediate neutrophilic progenitors, and the SB+ granule containing maturing neutrophils.32 To delineate at which stage neutrophil development is disrupted in c-mybhkz3/hkz3 mutants, we examined the expression of these different neutrophil markers and the staining of SB. Results showed that the cebp1+ early progenitors were indistinguishable between c-mybhkz3/hkz3 mutants and siblings (Figure 1G-H). Likewise, the mpx+ intermediate progenitors were formed properly in c-mybhkz3/hkz3 mutants, despite a slight decrease in number (Figure 1I-J). Meanwhile, the expression of cybb, which encodes the superoxide-generating enzyme in the neutrophil,47 is intact in c-mybhkz3/hkz3 mutants (Figure 1K-L). These observations indicate that in c-mybhkz3/hkz3 mutants the initiation of the neutrophilic program is intact and the majority of the early neutrophilic progenitors are capable of differentiating into the intermediate neutrophilic progenitors. These results also suggest that the absence of lyz expression in c-mybhkz3/hkz3 mutants is largely attributed to the downregulation of gene transcription, implying that c-Myb may govern neutrophil maturation by regulating the transcription of granule protein genes. Indeed, we found that the expression of serglycin (srgn), another granule proteoglycan protein highly enriched in neutrophils (supplemental Figure 1A-C, available on the Blood Web site),48 was also significantly downregulated in c-mybhkz3/hkz3 mutants (Figure 1M-N). The drastic decrease of srgn and lyz expression indicates that the granule formation in the c-mybhkz3/hkz3 mutant neutrophils is compromised. This notion was supported by SB-staining analysis, which showed that the SB+ neutrophils in 36 hpf c-mybhkz3/hkz3 embryos were reduced to 50% of sibling counts (Figure 1O-P), and more importantly, the signal intensity per cell for SB staining of the remaining neutrophils in the mutants was profoundly pale compared with those in siblings, suggesting that the c-mybhkz3/hkz3 neutrophils might have less abundant properly assembled granules. Indeed, VE DIC analysis of live embryos showed that the majority of the neutrophils identified in 2-dpf c-mybhkz3/hkz3 mutants possessed granules that were scant in their abundance and motility as opposed to the ample motile granule in siblings of the same stage (Figure 1Q-S). Furthermore, May-Grünwald-Giemsa staining showed a significant decrease of mature neutrophils with banded and segmented nuclei49,50 in 3-dpf c-mybhkz3/hkz3 mutants compared with that in siblings (Figure 1T). To further confirm that the neutrophil defects in c-mybhkz3/hkz3 mutants is caused by the loss of c-Myb function, we knocked down c-myb expression by morpholino43 and showed that the c-myb morpholino knockdown embryos displayed neutrophil defects similar to c-mybhkz3/hkz3 mutants (supplemental Figure 2A-H). Moreover, ectopically overexpression of c-Myb by the heat-shock–inducible promoter could restore the lyz expression and SB+ cell numbers in c-mybhkz3/hkz3 mutants (supplemental Figure 2I-R). From these results, we conclude that loss of c-Myb function blocks neutrophil differentiation, resulting in the accumulation of immature neutrophils with less segmented nuclei and scant granules, a defect similar to the SGD in mammals.14-16
c-Myb directly regulates the transcription of the granule-related gene lyz
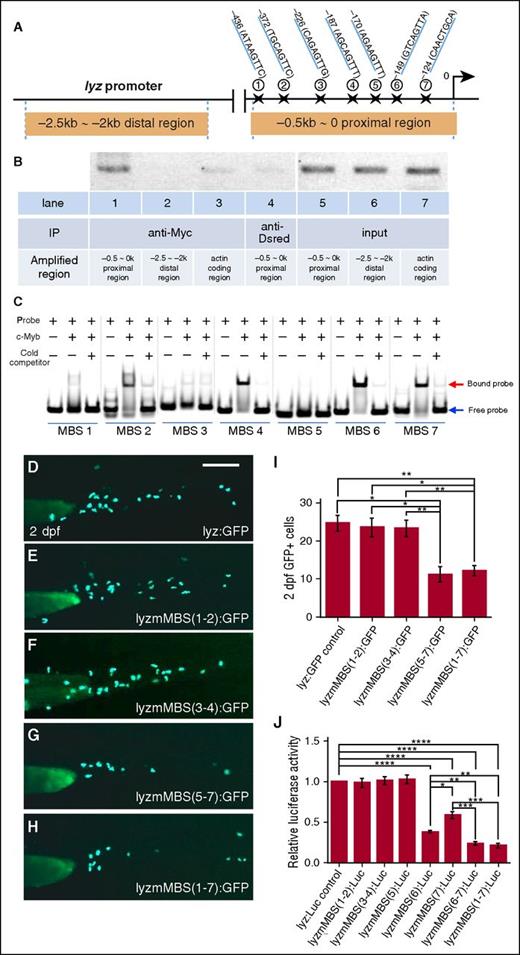
The blockage of neutrophil differentiation and the downregulation of several granule proteins in c-mybhkz3/hkz3 mutants (Figure 1) suggest that c-Myb function is required for the transcription of certain granule proteins. We therefore focused on lyz, the most widely expressed granule proteins found in all subtypes of neutrophil granules.44-46 To ascertain whether c-Myb directly regulates lyz expression, we first carried out bioinformatics analysis and identified 7 putative c-Myb–binding sites (MBSs) within the proximal promoter region (−0.5∼0 kb) of the lyz promoter (Figure 2A),51 raising the possibility that c-Myb might regulate lyz transcription by directly binding to the −0.5∼0 kb proximal promoter region. To support this notion, we performed chromatin immunoprecipitation (ChIP) assays using embryos injected with Myc-tagged c-myb mRNA to test whether recombinant Myc-tagged c-Myb proteins could bind the proximal promoter region of the lyz promoter. As shown in Figure 2B, pulling down recombinant c-Myb protein with anti-Myc antibody coprecipitated the −0.5∼0 kb proximal region of the lyz promoter, but not the −2.5 kb∼-2 kb distal region of the lyz promoter or 0∼0.5 kb within the coding region of actin gene, indicating that the −0.5∼0 kb proximal promoter region of the lyz promoter is occupied by c-Myb. To further determine which MBS is critical for c-Myb binding, we performed electrophoretic mobility shift assay (EMSA) and results showed that MBS-6 had the strongest binding ability with c-Myb, whereas MBS-4 and -7 exhibited moderate binding ability and the other MBSs (1, 2, 3, and 5) displayed weak or no binding ability (Figure 2C). In concordance with the electrophoretic mobility shift assay (EMSA) results, transient GFP reporter assay in zebrafish embryos and luciferase reporter assay in 293T cells showed that disruption of MBS-6 and MBS-7 dramatically reduced the lyz promoter activity as well (Figure 2D-J). Taken together, these data demonstrate that c-Myb regulates the lyz transcription by directly binding to its promoter region, especially MBS-6 and -7.
lyz is a direct downstream target of c-Myb. (A) Schematic diagram of the 2.5-kb lyz promoter region. The transcription initiation site is designated as 0. Seven putative c-Myb consensus sites are identified within −0.5∼0 kb proximal region using promo 3.0 online software and their positions (marked by stars) relative to the transcription initiation site are shown. (B) ChIP shows that c-Myb binds to the −0.5-kb proximal promoter region of the lyz promoter. Lysates from the embryos injected with the Myc-tagged c-myb mRNA were precipitated with anti-Myc (lanes 1-3) and anti-Dsred (lane 4, negative control) antibodies. The precipitates were then subjected to semiquantitative PCR analysis of the enrichment of the −0.5∼0-kb proximal region (lanes 1 and 4), the −2.5- ∼ −2-kb distal region (lane 2), and the actin-coding region (lane 3). Lanes 5-7 are input DNA control. (C) Gel shift image shows that 7 FAM-labeled probes containing putative MBSs were incubated with purified His-c-Myb N-terminal (amino acids 33-202) of the zebrafish c-Myb protein, encompassing the highly conserved DNA-binding domain. Excess unlabeled probes compete with the FAM-labeled probes. Equal amounts of proteins were used, specifically bound to the FAM-labeled probes. (D-I) Transient GFP reporter assay. Embryos were injected at the 1-cell stage with pTol-lyzPromoter-GFP reporter constructs driven by an intact 2.4-kb lyz promoter: pTol-lyz:GFP (D) or lyz promoter with subsets of the mutated c-MBSs (mMBS): pTol-lyzmMBS(1-2):GFP (E), pTol-lyzmMBS(3-4):GFP (F), pTol-lyzmMBS(5-7):GFP (G), and pTol-lyzmMBS(1-7):GFP (H). The promoter activity was then measured by calculating the numbers of GFP+ cells at 2 dpf. Representative images (D-H) and GFP+ quantifications (I) of 2-dpf injected embryos (ANOVA, Dunnett T3, n ≥ 50). Scale bars represent 200 μm. (J) Luciferase reporter assay. Bars showed the relative luciferase activity of lyz promoter with different subsets of MBS mutation compared with unmutated control. The statistical significance was calculated using 1-way ANOVA followed by Dunnett T3 correction. An asterisk indicates a statistical difference (*P < .05, **P < .01, ***P < .001, ****P < .0001, triplicated).
lyz is a direct downstream target of c-Myb. (A) Schematic diagram of the 2.5-kb lyz promoter region. The transcription initiation site is designated as 0. Seven putative c-Myb consensus sites are identified within −0.5∼0 kb proximal region using promo 3.0 online software and their positions (marked by stars) relative to the transcription initiation site are shown. (B) ChIP shows that c-Myb binds to the −0.5-kb proximal promoter region of the lyz promoter. Lysates from the embryos injected with the Myc-tagged c-myb mRNA were precipitated with anti-Myc (lanes 1-3) and anti-Dsred (lane 4, negative control) antibodies. The precipitates were then subjected to semiquantitative PCR analysis of the enrichment of the −0.5∼0-kb proximal region (lanes 1 and 4), the −2.5- ∼ −2-kb distal region (lane 2), and the actin-coding region (lane 3). Lanes 5-7 are input DNA control. (C) Gel shift image shows that 7 FAM-labeled probes containing putative MBSs were incubated with purified His-c-Myb N-terminal (amino acids 33-202) of the zebrafish c-Myb protein, encompassing the highly conserved DNA-binding domain. Excess unlabeled probes compete with the FAM-labeled probes. Equal amounts of proteins were used, specifically bound to the FAM-labeled probes. (D-I) Transient GFP reporter assay. Embryos were injected at the 1-cell stage with pTol-lyzPromoter-GFP reporter constructs driven by an intact 2.4-kb lyz promoter: pTol-lyz:GFP (D) or lyz promoter with subsets of the mutated c-MBSs (mMBS): pTol-lyzmMBS(1-2):GFP (E), pTol-lyzmMBS(3-4):GFP (F), pTol-lyzmMBS(5-7):GFP (G), and pTol-lyzmMBS(1-7):GFP (H). The promoter activity was then measured by calculating the numbers of GFP+ cells at 2 dpf. Representative images (D-H) and GFP+ quantifications (I) of 2-dpf injected embryos (ANOVA, Dunnett T3, n ≥ 50). Scale bars represent 200 μm. (J) Luciferase reporter assay. Bars showed the relative luciferase activity of lyz promoter with different subsets of MBS mutation compared with unmutated control. The statistical significance was calculated using 1-way ANOVA followed by Dunnett T3 correction. An asterisk indicates a statistical difference (*P < .05, **P < .01, ***P < .001, ****P < .0001, triplicated).
c-myb and cebp1 genetically interact in regulating neutrophil maturation
Previous studies have shown that, similar to the role of C/EBPε in neutrophil development in mammals,14-16,52,53 transient knockdown of cebp1 expression by morpholino in zebrafish leads to the reduction of granule protein expression including lyz, without affecting the formation of macrophages and neutrophils.51,54,55 These findings prompted us to speculate that, in zebrafish, Cebp1 and c-Myb may form a genetic network to regulate neutrophil development.
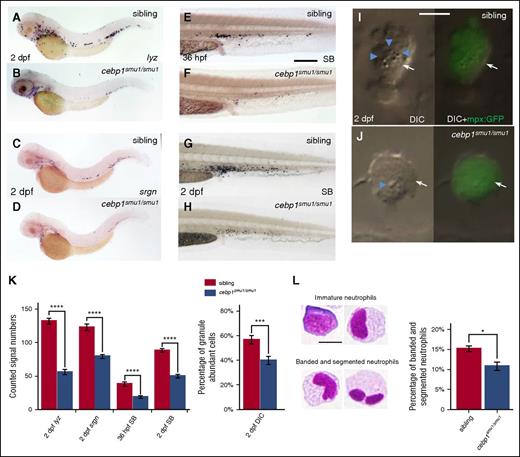
To unravel the genetic relationship of these 2 factors during zebrafish neutrophil development, we generated 2 frameshift cebp1 mutant fish by transcription activator-like effector nucleases (TALENs; cebp1smu1 and cebp1smu2) (supplemental Figure 3A-B) and asked whether inactivating Cebp1 would lead to the impairment of neutrophil development. We found that, although myeloid progenitors and macrophages remained unaffected (supplemental Figure 3C-H), the lyz+ and srgn+ neutrophils were clearly reduced in 2-dpf cebp1smu1/smu1 mutants (Figure 3A-D). The reduction of lyz and srgn staining in 2-dpf cebp1smu1/smu1 mutants was likely due to the decrease of lyz and srgn expression because the mpx+ neutrophils in 2-dpf cebp1smu1/smu1 mutants were comparable to those in control siblings (supplemental Figure 3I-J). As expected, the development of neutrophils in cebp1smu1/smu1 mutants was compromised as indicated by the accumulation of neutrophils with less abundant granules and unsegmented nuclei in cebp1smu1/smu1 mutants (Figure 3E-L). Likewise, the neutrophil development in adult cebp1smu1/smu1 mutants as indicated by the decreased expression of neutrophil markers in the kidney marrow cells was also impaired, whereas macrophages developed normally (supplemental Figure 3K). The other allele of cebp1 mutant, cebp1smu2, exhibited similar lyz+ reduction phenotype as cebp1smu1/smu1 (supplemental Figure 3L-M). These data reveal that, similar to the function of mammalian C/EBPε,56-58 Cebp1 plays a critical role in neutrophil differentiation in zebrafish.
Neutrophil maturation is attenuated in cebp1smu1/smu1 mutants. (A-B) WISH of lyz expression in 2-dpf siblings and cebp1smu1/smu1 mutants. (C-D) WISH of srgn expression in 2-dpf siblings and cebp1smu1/smu1 mutants. (E-H) SB staining of 36-hpf (E-F) and 2-dpf (G-H) siblings and cebp1smu1/smu1 mutants. (I-J) In vivo VE DIC microscopy reveals scant granule in neutrophils in 2 dpf Tg(mpx:GFP);cebp1smu1/smu1 transgenic mutants (J) compared with siblings (I). Left panels, Bright-field DIC images; right panels, overlays of bright-field DIC images with corresponding GFP fluorescent images. Blue arrowheads identify granules contained by neutrophils (white arrows) expressing GFP. (K) Quantifications of WISH, SB staining, and DIC analysis (Student t test, ***P < .001, ****P < .0001; WISH and SB, n ≥ 23; DIC, n ≥ 56). (L) May-Grünwald-Giemsa staining of neutrophils in 3-dpf embryos (left) and neutrophils were quantitated by morphology (right) (Student t test, *P < .05, n ≥ 500). Scale bars, 200 μm (E-H) and 5 μm (I, J, L).
Neutrophil maturation is attenuated in cebp1smu1/smu1 mutants. (A-B) WISH of lyz expression in 2-dpf siblings and cebp1smu1/smu1 mutants. (C-D) WISH of srgn expression in 2-dpf siblings and cebp1smu1/smu1 mutants. (E-H) SB staining of 36-hpf (E-F) and 2-dpf (G-H) siblings and cebp1smu1/smu1 mutants. (I-J) In vivo VE DIC microscopy reveals scant granule in neutrophils in 2 dpf Tg(mpx:GFP);cebp1smu1/smu1 transgenic mutants (J) compared with siblings (I). Left panels, Bright-field DIC images; right panels, overlays of bright-field DIC images with corresponding GFP fluorescent images. Blue arrowheads identify granules contained by neutrophils (white arrows) expressing GFP. (K) Quantifications of WISH, SB staining, and DIC analysis (Student t test, ***P < .001, ****P < .0001; WISH and SB, n ≥ 23; DIC, n ≥ 56). (L) May-Grünwald-Giemsa staining of neutrophils in 3-dpf embryos (left) and neutrophils were quantitated by morphology (right) (Student t test, *P < .05, n ≥ 500). Scale bars, 200 μm (E-H) and 5 μm (I, J, L).
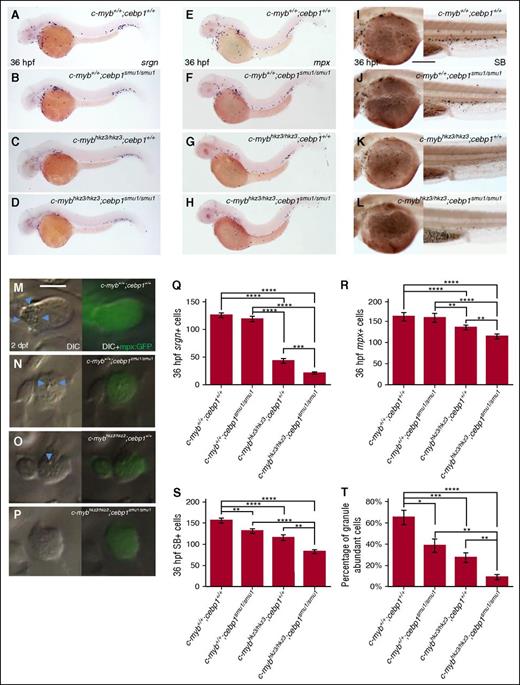
To delineate the genetic relationship of c-Myb and Cebp1 during neutrophil differentiation, we first investigated whether there was a mutual dependency of the expression of c-myb and cebp1 at around 22 hpf when embryonic neutrophils begun to undergo differentiation. Results showed that c-myb expression was unchanged in cebp1smu1/smu1 mutant embryos (supplemental Figure 3N-P), and likewise, cebp1 expression was also unaltered in c-mybhkz3/hkz3 embryos (Figure 1G-H). The independence of c-myb and cebp1 transcriptional regulation suggests that they may act in parallel and perhaps cooperatively to regulate neutrophil differentiation. To test this possibility, we generated c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants and asked whether inactivating both c-Myb and Cebp1 spontaneously would lead to a more severe neutrophil phenotype. Indeed, c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants displayed a much greater reduction of srgn, mpx, and SB staining compared with c-mybhkz3/hkz3;cebp1+/+ or c-myb+/+;cebp1smu1/smu1 single mutants (Figure 4A-D,E-H,I-L,Q-S). Moreover, the percentage of immature neutrophils in c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants was significantly increased compared with c-mybhkz3/hkz3;cebp1+/+ or c-myb+/+;cebp1smu1/smu1 single mutants (Figure 4M-P,T). Collectively, these genetic data suggest that c-Myb and Cebp1 act in parallel and cooperatively to regulate neutrophil terminal maturation.
c-myb and cebp1 genetically interact in regulating neutrophil maturation. (A-D) WISH shows further decrease of srgn expression in 36-hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (D) compared with c-myb+/+;cebp1smu1/smu1 (B) or c-mybhkz3/hkz3;cebp1+/+ (C) single mutants. (E-H) WISH shows further decrease of mpx expression in 36-hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (H) compared with c-myb+/+;cebp1smu1/smu1 (F) or c-mybhkz3/hkz3;cebp1+/+ (G) single mutants. (I-L) SB staining shows further decrease of SB+ cells in 36 hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (L) compared with c-myb+/+;cebp1smu1/smu1 (J) or c-mybhkz3/hkz3;cebp1+/+ (K) single mutants. (M-P) In vivo VE DIC microscopy reveals further reduction of granules in neutrophils in 2-dpf Tg(mpx:GFP);c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (P) compared with Tg(mpx:GFP);c-myb+/+;cebp1smu1/smu1 (N) or Tg(mpx:GFP);c-mybhkz3/hkz3;cebp1+/+ (O) single mutants. Left panels, Bright-field DIC images. Right panels, Overlays of bright-field DIC images with corresponding GFP fluorescent images. Blue arrowheads indicate granules contained by neutrophils. (Q) Quantifications of srgn+ cells in 36 hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double and single mutants. No statistical difference is found between c-myb+/+;cebp1+/+ and c-myb+/+;cebp1smu1/smu1. An asterisk indicates a statistical difference (ANOVA, Dunnett T3, n ≥ 16). (R) Quantifications of mpx+ cells in 36-hpf WT, c-mybhkz3/hkz3;cebp1smu1/smu1double and single mutants. No statistical difference is found between c-myb+/+;cebp1+/+ and c-myb+/+;cebp1smu1/smu1. An asterisk indicates a statistical difference (ANOVA, Dunnett T3, n ≥ 20). (S) Quantifications of SB+ cells in 36-hpf WT, c-mybhkz3/hkz3;cebp1smu1/smu1 double and single mutants. An asterisk indicates a statistical difference (ANOVA, Bonferroni, n ≥ 18). (T) Percentage of neutrophils with abundant granules in 2-dpf WT, c-mybhkz3/hkz3;cebp1smu1/smu1 double and single mutants (ANOVA, Dunnett T3, n ≥ 16). An asterisk indicates a statistical difference (*P < .05, **P < .01, ***P < .001, ****P < .0001). Scale bars, 200 μm (I-L) and 5 μm (M-P).
c-myb and cebp1 genetically interact in regulating neutrophil maturation. (A-D) WISH shows further decrease of srgn expression in 36-hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (D) compared with c-myb+/+;cebp1smu1/smu1 (B) or c-mybhkz3/hkz3;cebp1+/+ (C) single mutants. (E-H) WISH shows further decrease of mpx expression in 36-hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (H) compared with c-myb+/+;cebp1smu1/smu1 (F) or c-mybhkz3/hkz3;cebp1+/+ (G) single mutants. (I-L) SB staining shows further decrease of SB+ cells in 36 hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (L) compared with c-myb+/+;cebp1smu1/smu1 (J) or c-mybhkz3/hkz3;cebp1+/+ (K) single mutants. (M-P) In vivo VE DIC microscopy reveals further reduction of granules in neutrophils in 2-dpf Tg(mpx:GFP);c-mybhkz3/hkz3;cebp1smu1/smu1 double mutants (P) compared with Tg(mpx:GFP);c-myb+/+;cebp1smu1/smu1 (N) or Tg(mpx:GFP);c-mybhkz3/hkz3;cebp1+/+ (O) single mutants. Left panels, Bright-field DIC images. Right panels, Overlays of bright-field DIC images with corresponding GFP fluorescent images. Blue arrowheads indicate granules contained by neutrophils. (Q) Quantifications of srgn+ cells in 36 hpf c-mybhkz3/hkz3;cebp1smu1/smu1 double and single mutants. No statistical difference is found between c-myb+/+;cebp1+/+ and c-myb+/+;cebp1smu1/smu1. An asterisk indicates a statistical difference (ANOVA, Dunnett T3, n ≥ 16). (R) Quantifications of mpx+ cells in 36-hpf WT, c-mybhkz3/hkz3;cebp1smu1/smu1double and single mutants. No statistical difference is found between c-myb+/+;cebp1+/+ and c-myb+/+;cebp1smu1/smu1. An asterisk indicates a statistical difference (ANOVA, Dunnett T3, n ≥ 20). (S) Quantifications of SB+ cells in 36-hpf WT, c-mybhkz3/hkz3;cebp1smu1/smu1 double and single mutants. An asterisk indicates a statistical difference (ANOVA, Bonferroni, n ≥ 18). (T) Percentage of neutrophils with abundant granules in 2-dpf WT, c-mybhkz3/hkz3;cebp1smu1/smu1 double and single mutants (ANOVA, Dunnett T3, n ≥ 16). An asterisk indicates a statistical difference (*P < .05, **P < .01, ***P < .001, ****P < .0001). Scale bars, 200 μm (I-L) and 5 μm (M-P).
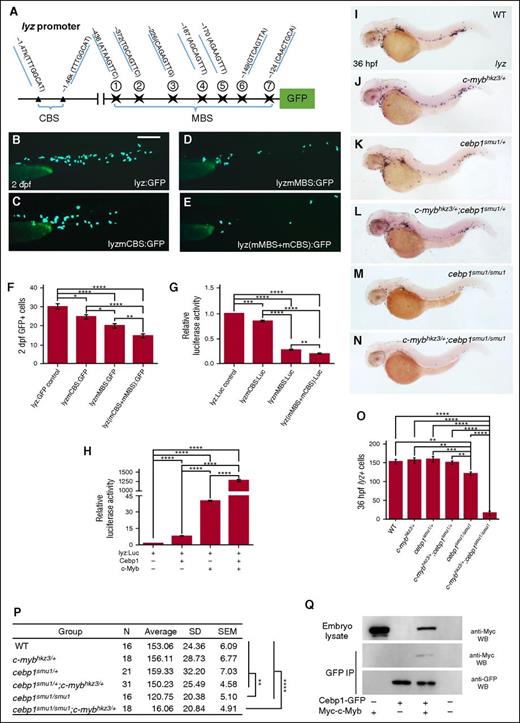
c-Myb and Cebp1 synergistically regulate lyz transcription activation
The genetic interaction of c-Myb and Cebp1 led us to hypothesize that the 2 factors govern neutrophil differentiation by coregulating the expression of downstream target genes, especially the genes associated with granule formation. To support this idea, we again focused on the transcriptional regulation of lyz, a key granule protein known to be regulated by both c-Myb (Figure 2) and Cebp1.51 Previous study has shown that a 2.4-kb DNA fragment upstream of the lyz transcription initiation site is sufficient to direct GFP reporter expression in a manner similar to that of the endogenous lyz,51 indicating that this 2.4-kb DNA fragment likely contains the critical cis elements for the regulation of lyz expression. Indeed, a cluster of putative MBSs and another cluster of Cebp1-binding sites (CBSs) were found within this 2.4-kb fragment (Figure 5A) and these clusters have been shown to be recognized by c-Myb (Figure 2) and Cebp1,51 respectively. To examine the cooperative effect of c-Myb and Cebp1 on lyz transcription, we injected wild-type (WT) 2.4-kb lyz promoter reporter construct and the mutated 2.4-kb lyz promoter reporter construct into zebrafish embryos and assayed the promoter activity. Our results showed that the disruption of either MBS or CBS alone reduced the activity of lyz promoter as indicated by the decrease of GFP expression in the myeloid cells in the embryos injected with respective construct (Figure 5B-D,F). Consistent with the genetic analysis, when MBS and CBS were mutated simultaneously, GFP expression was further reduced (Figure 5E-F), indicating that the binding of c-Myb and Cebp1 to the lyz promoter contributes to the activation of lyz transcription. A similar conclusion was obtained with lyz promoter luciferase reporter assay (Figure 5G). These data prompted us to speculate that c-Myb and Cebp1 might regulate lyz transcription synergistically. To support this notion, we compared the lyz promoter activity in 293T cells overexpressing c-Myb and Cebp1 individually or simultaneously. As anticipated, results showed that, when c-Myb and Cebp1 were cotransfected, the luciferase activity was much higher than the combined values with c-Myb or Cebp1 alone (Figure 5H), indicating that the 2 transcription factors act synergistically to regulate lyz transcription.
c-Myb and Cebp1 synergistically regulate lyz transcription. (A) Schematic diagram of lyz promoter region. The transcription initiation site is designated as 0. Seven putative c-Myb (marked by star) and 2 Cebp1 (marked by triangle) consensus sites are identified using promo 3.0 online software and their positions relative to the transcription initiation site are shown. (B-E) Transient reporter assay demonstrates that c-Myb and Cebp1 synergistically regulate lyz transcription. Embryos were injected at 1-cell stage with pTol-GFP reporter constructs driven by an intact 2.4-kb lyz promoter (lyz:GFP), c-Myb sites mutated lyz promoter pTol-lyz(mMBS):GFP, Cebp1 sites mutated lyz promoter pTol-lyz(mCBS):GFP or both sites mutated pTol-lyz(mMBS+mCBS):GFP, and lyz promoter activity were measured at 2 dpf by scoring the number of GFP+ cells. Scale bars represent 200 μm. (F) Quantification of GFP+ cells in 2 dpf pTol-lyz:GFP, pTol-lyz(mCBS):GFP, pTol-lyz(mMBS):GFP, and pTol-lyz(mCBS+mMBS):GFP-injected embryos (ANOVA, Dunnett T3, n ≥ 160). (G-H) Luciferase reporter assay. Bars showed the relative luciferase activity of transfection with lyz(mMBS):Luc, lyzCBS(mCBS):Luc or both sites mutated lyz(mMBS+mCBS):Luc compared with unmutated lyz:Luc control (G). Synergistic effect on transactivation of lyz promoter by c-Myb and Cebp1 (H). Bars showed the relative luciferase activity with overexpression of cebp1, c-myb, or both genes compared with control (H). The statistical significance was calculated using 1-way ANOVA followed by Dunnett T3 correction. The asterisk indicates a statistical difference (*P < .05, **P < .01, ***P < .001, ****P < .0001, triplicated). (I-N) WISH shows that lyz expression is further decreased in 36-hpf c-mybhkz3/+;cebp1smu1/smu1 double (cebp1 homozygous and cmyb heterozygous) mutants (N) compared with cebp1smu1/smu1 single mutants (M). (O-P) Quantifications of lyz+ cells in 36-hpf WT (c-myb+/+;cebp1+/+), single-heterozygous (c-mybhkz3/+ and cebp1smu1/+), double-heterozygous (c-mybhkz3/+;cebp1smu1/+) mutant embryos, cebp1smu1/smu1 single mutants and c-mybhkz3/+;cebp1smu1/smu1 double mutants shown in the statistical figure (O) and table (P). The statistical significance was calculated using 1-way ANOVA followed by Bonferroni correction. The asterisk indicates a statistical difference (**P < .01, ***P < .001, ****P < .0001, n ≥ 16). (Q) Coimmunoprecipitation experiment shows a physical interaction between c-Myb and Cebp1. Embryos overexpressed with Myc-tagged c-myb, and GFP-tagged cebp1 by heat-shock induction were immunoprecipitated with anti-GFP antibody. The immunoprecipitates were subjected to western blotting with anti-Myc and anti-GFP antibody.
c-Myb and Cebp1 synergistically regulate lyz transcription. (A) Schematic diagram of lyz promoter region. The transcription initiation site is designated as 0. Seven putative c-Myb (marked by star) and 2 Cebp1 (marked by triangle) consensus sites are identified using promo 3.0 online software and their positions relative to the transcription initiation site are shown. (B-E) Transient reporter assay demonstrates that c-Myb and Cebp1 synergistically regulate lyz transcription. Embryos were injected at 1-cell stage with pTol-GFP reporter constructs driven by an intact 2.4-kb lyz promoter (lyz:GFP), c-Myb sites mutated lyz promoter pTol-lyz(mMBS):GFP, Cebp1 sites mutated lyz promoter pTol-lyz(mCBS):GFP or both sites mutated pTol-lyz(mMBS+mCBS):GFP, and lyz promoter activity were measured at 2 dpf by scoring the number of GFP+ cells. Scale bars represent 200 μm. (F) Quantification of GFP+ cells in 2 dpf pTol-lyz:GFP, pTol-lyz(mCBS):GFP, pTol-lyz(mMBS):GFP, and pTol-lyz(mCBS+mMBS):GFP-injected embryos (ANOVA, Dunnett T3, n ≥ 160). (G-H) Luciferase reporter assay. Bars showed the relative luciferase activity of transfection with lyz(mMBS):Luc, lyzCBS(mCBS):Luc or both sites mutated lyz(mMBS+mCBS):Luc compared with unmutated lyz:Luc control (G). Synergistic effect on transactivation of lyz promoter by c-Myb and Cebp1 (H). Bars showed the relative luciferase activity with overexpression of cebp1, c-myb, or both genes compared with control (H). The statistical significance was calculated using 1-way ANOVA followed by Dunnett T3 correction. The asterisk indicates a statistical difference (*P < .05, **P < .01, ***P < .001, ****P < .0001, triplicated). (I-N) WISH shows that lyz expression is further decreased in 36-hpf c-mybhkz3/+;cebp1smu1/smu1 double (cebp1 homozygous and cmyb heterozygous) mutants (N) compared with cebp1smu1/smu1 single mutants (M). (O-P) Quantifications of lyz+ cells in 36-hpf WT (c-myb+/+;cebp1+/+), single-heterozygous (c-mybhkz3/+ and cebp1smu1/+), double-heterozygous (c-mybhkz3/+;cebp1smu1/+) mutant embryos, cebp1smu1/smu1 single mutants and c-mybhkz3/+;cebp1smu1/smu1 double mutants shown in the statistical figure (O) and table (P). The statistical significance was calculated using 1-way ANOVA followed by Bonferroni correction. The asterisk indicates a statistical difference (**P < .01, ***P < .001, ****P < .0001, n ≥ 16). (Q) Coimmunoprecipitation experiment shows a physical interaction between c-Myb and Cebp1. Embryos overexpressed with Myc-tagged c-myb, and GFP-tagged cebp1 by heat-shock induction were immunoprecipitated with anti-GFP antibody. The immunoprecipitates were subjected to western blotting with anti-Myc and anti-GFP antibody.
To further support the results obtained from lyz promoter reporter assay, we generated a combination of double mutants expressing different levels of c-Myb and Cebp1 and subsequently profiled lyz expression at 36 hpf. c-myb+/+;cebp1+/+, c-mybhkz3/+;cebp1+/+, c-myb+/+;cebp1smu1/+, c-mybhkz3/+;cebp1smu1/+, c-myb+/+;cebp1smu1/smu1, and c-mybhkz3/+;cebp1smu1/smu1 embryos expressed different levels of c-Myb and Cebp1 and profiled lyz expression at 36 hpf. As shown in Figure 5I-P, single-heterozygous (c-mybhkz3/+;cebp1+/+ and c-myb+/+;cebp1smu1/+) and double-heterozygous (c-mybhkz3/+;cebp1smu1/+) mutant embryos had comparable lyz staining with the WT (c-myb+/+;cebp1+/+) embryos (Figure 5I-L,O-P), whereas cebp1 single-homozygous (c-myb+/+;cebp1smu1/smu1) displayed a slight reduction in lyz staining (reduced by ∼20%) (Figure 5M,O-P) (c-myb single-homozygous c-mybhkz3/hkz3 contained a very low level of lyz expression and was hardly detectable by WISH; Figure 1B). However, as we expected, a dramatic decrease of lyz expression (reduced by ∼90%) was seen in the cebp1-homozygous plus c-myb-heterozygous (c-mybhkz3/+;cebp1smu1/smu1) double-mutant embryos compared with the cebp1 single-homozygous (c-myb+/+;cebp1smu1/smu1) mutant embryos (Figure 5M-P), showing that the suppression of lyz expression in cebp1smu1/smu1 was further exacerbated by c-mybhkz3/+ heterozygosity.
The cooperative effect of c-Myb and Cebp1 on the lyz promoter suggests that the 2 factors might be part of the activation complexes critical for lyz transcription. To test this hypothesis, we performed a coimmunoprecipitation experiment in transgenic zebrafish embryos with heat-shock–induced Cebp1-GFP and Myc-c-Myb. Immunoprecipitating GFP-tagged Cebp1 with an anti-GFP antibody indeed resulted in coprecipitation of the Myc-tagged c-Myb (Figure 5Q), suggesting that Cebp1 and c-Myb are physically interacted, directly or indirectly, to regulate lyz transcription in a cooperative manner. Taken together, the genetic epistasis and biochemical analysis conclude that c-Myb and Cebp1 act in parallel and cooperatively to regulate neutrophil maturation by synergistically regulating granule protein gene transcription.
Discussion
In this study, we uncover transcription factor c-Myb as a new molecular determinant involved in neutrophil terminal differentiation during zebrafish embryonic myelopoiesis. Since discovered as the cellular counterpart of v-MYB, an oncogene presented in 2 avian retroviruses associated with myeloid leukemia,18 c-MYB was thought to have a critical role in myeloid lineage development. Despite a number of genes found to be regulated by c-MYB and overexpression of c-MYB blocking monocyte/macrophage differentiation in cultural cell lines,59,60 in vivo knockout studies in mice have revealed that c-MYB is mainly involved in the development of HSCs and the formation of eosinophils.24,61 Whether c-MYB has a role in the regulation of the development of 2 main myeloid lineages, macrophages, and neutrophils, remains uncertain. Our present study provides evidence demonstrating that c-Myb is essential for terminal differentiation, especially granule formation, of neutrophilic granulocytes during zebrafish embryonic myelopoiesis. Intriguingly, inactivation of c-Myb appears to have little impact on the development of myeloid progenitors and macrophages, despite its enriched expression in these cells. Why c-Myb is required for neutrophil differentiation but is dispensable for myeloid progenitor and macrophage lineage development remains completely unknown; further in-depth analysis will be required to clarify this issue. Another interesting question, which cannot be addressed in current study because of the block of adult hematopoiesis at the HSC level in c-mybhkz3/hkz3 mutants,37 is whether c-Myb has a similar role during adult myelopoiesis. Generation of a myeloid-specific c-myb–deficient mutant fish will be necessary to answer this question. Finally, our findings prompt us to speculate that perhaps mammalian c-MYB also plays a similar role in neutrophil differentiation. Thus, it will be of great interest to reinvestigate the role of c-MYB in murine embryonic and adult myelopoiesis.
Our study also shows that the neutrophilic phenotype in c-mybhkz3/hkz3 mutants is largely attributed to the downregulation of differentiation-related genes such as lyz and srgn, the key components of granule proteins.44-46,48 This result is somewhat surprising in view of a well-documented role of c-MYB in cell-cycle regulation.62 It may reflect a disparate requirement for c-Myb in distinct cellular contexts, which is also supported by our previous findings showing that c-Myb regulates the migration of newly born HSCs by modulating the expression of sdf1a.37 Consistent with previous in vitro cell line studies showing that c-MYB could directly interact with C/EBPε and cooperatively activate transcription of several myeloid-specific promoters (such as elastase, mim-1, and mpo),63-67 our genetic and biochemistry analysis indicates that c-Myb interacts with Cebp1, but they act in parallel and cooperatively to regulate neutrophil terminal differentiation, presumably through coactivating the granule-forming related gene transcription. Intriguingly, the loss of c-Myb function appears to have more profound impacts on neutrophil development than the loss of Cebp1 function, suggesting that c-Myb plays a more important role in the neutrophil maturation pathway. Besides the synergistic effect of c-Myb and C/ebp1 we reported here, other combined effects on lyz transcriptional regulation by different transcription factors (such as Runx151 together with c-Myb or Cebp1) remain currently unknown and need future study.
Finally, the neutrophilic defect in c-mybhkz3/hkz3 mutants is reminiscent of the pathological features of SGD, which is characterized by recurrent bacterial infections due to disabled neutrophils. The neutrophils in SGD patients display typical bilobed nuclei and lack the expression of at least 1 primary granule or/and all secondary and tertiary granule proteins.16 To date, the mutations in C/EBPε are the only known genetic lesions associated with SGD, but several classic SGD cases are shown to have intact C/EBPε.17 Our study strongly suggests that c-MYB could be an unidentified gene mutated in those SGD patients with intact C/EBPε alleles and it would be of great interest to investigate whether these human patients carry mutations in the c-MYB locus.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Nathan Lawson and Koichi Kawakami, Philip W. Ingham and Steve Renshaw, Philippe Herbomel and Bo Zhang for providing the pTol vector, the Tg(mpx:eGFP) line, DIC system advice, and TALEN reagents and protocol. The authors thank Laixin Xia, Kuangyu Yen, Qiang Wang, Shan Xiao, Zhaoting Liu, Fang Luo, Jingjing Zhang, Qi Xie, and Qian Chen for their helpful suggestions.
This work was supported by the National Natural Science Foundation of China (31271574; 31229003), the Research Grants Council of the Hong Kong Special Administrative Region (HKSAR) (663212; HKUST5/CRF/12R; AoE/M-09/12), the Innovation and Technology Commission of the HKSAR (ITCPD/17-9), and the Team Program of Guangdong Natural Science Foundation (2014A030312002).
Authorship
Contribution: H.J., Y.Z., Z.W., W.Z., and L.L. designed the experiments; H.J. and Z.H. designed and performed experiments; M.W., Y.C., and Y.L. performed the reporter assay; M.W. performed the rescue assay and generated the cebp1smu2 mutant; L.Z. generated the cebp1smu1 mutant; R.Z. performed blood cell staining and generated constructs; Y.Z., Z.H., and M.W. performed biochemistry experiments; L.L. characterized the c-mybhkz3 mutant; H.J., Z.H., Y.Z., and Z.W. wrote the manuscript; and J.X., F.Z., L.L., W.Z., Z.W., and Y.Z. discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.J. is Howard Hughes Medical Institute, Columbia College of Physicians and Surgeons, Columbia University, New York, NY.
Correspondence: Zilong Wen, State Key Laboratory of Molecular Neuroscience, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, People’s Republic of China; e-mail: zilong@ust.hk; and Yiyue Zhang, Department of Developmental Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, People’s Republic of China; e-mail: yiyue@smu.edu.cn.
References
Author notes
H.J. and Z.H. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal