Abstract
Over the past decade, there have been numerous published candidate gene association studies of non-HLA variants, genes and/or pathways in relation to BMT survival outcomes. Most of these studies investigated a small number of candidate single nucleotide polymorphisms (SNPs), chosen based on known gene function, in a limited number of cases without replication. Here we test the association of previously reported SNPs with survival outcomes (overall survival (OS), progression-free survival (PFS), death due to disease (DD) or transplant-related mortality (TRM)) after URD BMT using data from the DISCOVeRY-BMT (Determining the Influence of Susceptibility Conveying Variants Related to one-Year mortality after BMT) study which included 2,052 European descent recipient-unrelated donor pairs (Cohort 1) and 763 recipient-unrelated donor pairs (Cohort 1) reported to the Center for International Blood and Marrow Transplant Research (CIBMTR) between 2000-2009 and 2009-2011, respectively. DISCOVeRY-BMT is a Genome Wide Association Study (GWAS) that includes over 9 million typed or imputed SNPs. This represents the largest study to date and is well powered to test both SNP and gene-based association hypotheses.
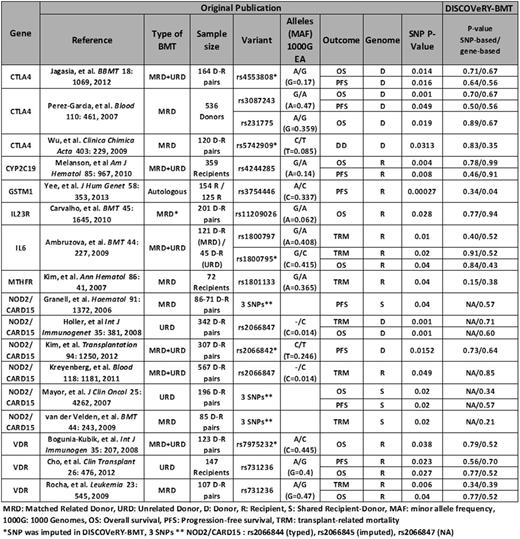
We conducted an extensive literature review of PubMed using MESH terms related to transplant and hematologic disease which yielded 76 candidate gene association studies that investigated a total of 46 genes with any survival outcome. Of the 46 genes, 11 genes (CTLA4, CYP2B6, CYP2C19, GSTM1, GSTT1, IL23R, IL6, MTHFR, NOD2/CARD15, TNFSRF1B, and VDR) were studied in at least 2 publications. A total of 16 SNPs in 8 of these genes reported a significant (P< 0.05) association with at least one survival outcome (OS, PFS, DD or TRM). We tested these 16 SNPs for an association with the survival outcome originally reported in each article (Table 1). All association models included age at BMT, diagnosis (AML, ALL, MDS), disease status at BMT, cell source (blood, bone marrow) and year of BMT. P-values from Cohorts 1 and 2 were combined using a fixed-effects model in METAL software. Since the candidate gene studies have a gene-based hypothesis, we also performed gene-level testing using the VErsatile Gene-Based Association Study (VEGAS2) software to measure the aggregate effect of all available typed and imputed SNPs from our GWAS data in these 8 genes.
The candidate gene association studies included heterogeneous patient populations of autologous and/or allogeneic (related or unrelated donors) BMT for a variety of hematologic diseases. Most candidate gene studies performed genotyping on both donors and recipients, and SNPs were tested either independently for donor or recipient genomes, or considering the total number of variant alleles the recipient-donor pair had at a given locus (0-4) (Table 1).
The 16 SNPs from the previous candidate gene association studies were not significantly associated at a nominal P<0.05 to either the survival outcome reported in the original published paper, or to any survival outcome (Table 1). Likewise, the VEGAS2 gene-based analysis did not reveal any statistically significant associations for any of the 8 genes at P<0.05. Our study also corroborated prior reports of SNPs and genes that were not associated with survival outcomes.
Candidate gene association studies use a priori biological knowledge to select genes that are likely associated with outcome and subsequently SNPs within the gene thought likely to affect function based on knowledge at the time of the investigation. Our findings suggest that neither the genetic variation tested in these candidate genes, nor other individual SNPs in the gene independently or collectively are not associated with survival outcomes in recipients of URD BMT. Our failure to replicate the prior candidate gene studies may be due to these SNPs being dependent on the underlying disease (ie. AML, ALL, etc), the donor relation (related, unrelated, autologous), or that there is no true association of these SNPs with the outcomes studied.
#These authors contributed equally
McCarthy:Gamida Cell: Honoraria, Membership on an entity's Board of Directors or advisory committees; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Karyopharm: Honoraria, Membership on an entity's Board of Directors or advisory committees; Onyx: Honoraria, Membership on an entity's Board of Directors or advisory committees; Bristol Myers Squibb: Honoraria, Membership on an entity's Board of Directors or advisory committees; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees; The Binding Site: Honoraria, Membership on an entity's Board of Directors or advisory committees; Sanofi: Honoraria, Membership on an entity's Board of Directors or advisory committees. Hahn:Novartis: Equity Ownership; NIH: Research Funding. Sucheston-Campbell:NIH/NCI: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal