Abstract
Mixed Lineage Leukemia gene rearrangements (MLL-r) account for nearly 10% of human acute leukemia cases and are generally associated with poor prognosis. Previous studies have revealed an essential role of the histone H3K79 methyltransferase Disruptor of Telomeric Silencing-1 Like (DOT1L) in MLL-r leukemogenesis. Our recent report (Chen et al. 2015 Nature Medicine) further identified a role for histone acetylation in DOT1L dependent gene expression driven by MLL-fusion proteins including MEIS1 and HOXA cluster genes. A first-in-human Phase I clinical trial demonstrated clinical activity of DOT1L inhibition in MLL-r leukemia patients, thus providing a potential opportunity for treating these malignant diseases. Nevertheless, the incomplete silencing of the leukemic program by only targeting DOT1L motivates the need for additional and perhaps combinational approaches to improve therapies against MLL-r leukemias.
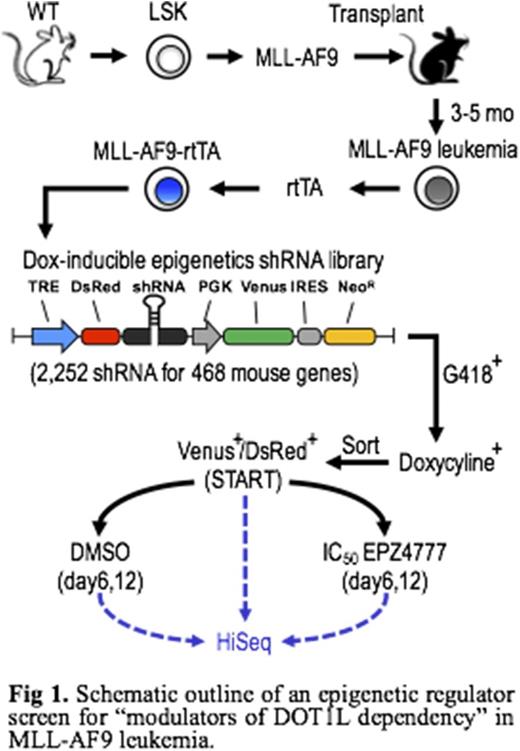
To enhance the efficacy of DOT1L inhibition, we sought to identify genes whose suppression would synergize with the DOT1L inhibitors to suppress the proliferation of mouse bone marrow progenitors transformed with MLL-AF9. We conducted a pooled RNAi screen using a customized library composed of 2,252 shRNA targeting 468 epigenetic regulators (i.e. writers, readers, and erasers of chromatin modifications; Fig 1). The integrated shRNA sequences were assessed using high-throughput sequencing. By comparing the change in frequency of each shRNA construct cultured in control vs. an IC50 DOT1L inhibitor EPZ4777, we identified several candidate modulators of DOT1L dependency, which had multiple shRNAs selectivity depleted only in the DOT1L suppressed condition. Notably, using a network correlation study, we found that one of the top candidate genes Plant Homeodomain Finger Protein 20 (PHF20) is highly associated with histone acetylation in the mammalian epigenome. Knockdown of PHF20 drastically increased the sensitivity of MLL-AF9 leukemic blasts to DOT1L inhibitors through enhanced myeloid differentiation and reduced cell proliferation, colony formation, and re-plating capacity. Similar phenotypes were also observed in PHF20-deficient MLL-AF9 cells generated by CRISPR/Cas9-mediated gene knockout.
PHF20 is an epigenetic adaptor protein that has no predicted enzymatic activity. To investigate the role of PHF20, we conducted a CRISPR functional domain screen and identified the requirement of the chromatin reader domains in PHF20, including the Tudor domains and the PHD-finger, in supporting the survival of MLL-r leukemic cells upon DOT1L inhibition. We also performed RNA-seq and found that suppression of PHF20 facilitated the silencing of the MLL-AF9 leukemic program induced by DOT1L inhibitor treatment. Chromatin immunoprecipitation and sequencing (ChIP-seq) analyses validated that PHF20 contributes to the maintenance of histone acetylation including H3K9ac and H4K16ac at MLL-AF9 target loci. In line with the profound loss of histone acetylation at MLL-AF9 target loci in PHF20-depleted cells, we found that knockdown of a known PHF20 interacting partner KAT8 (a histone acetyltransferase; also known as MOF or MYST1) phenocopies the effects observed in PHF20-knockdown cells. Finally, we showed that pharmacological inhibition of DOT1L and KAT8 synergistically suppresses the proliferation and survival of MLL-AF9 leukemic cells. These data collectively highlight the involvement of a novel DOT1L-PHF20-KAT8 axis in mammalian gene regulation and MLL-r leukemogenesis.
In summary, our studies show that MLL-rearrangements may drive leukemic transformation by coordinating an epigenetic network involving several histone modifications associated with gene transcription (e.g. H3K79 methylation and H3K9/H4K16 acetylation). Our results also suggest that simultaneous targeting of multiple components of this epigenetic feed-forward loop including DOT1L and PHF20/KAT8 may provide a novel and more effective approach against MLL-r leukemia.
Bradner:Novartis Institutes for BioMedical Research: Employment. Armstrong:Epizyme, Inc: Consultancy; Vitae Pharmaceuticals: Consultancy; Imago Biosciences: Consultancy; Janssen Pharmaceutical: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal