Abstract
Background: Whole genome amplification (WGA) has become an invaluable method for working with small amounts of starting DNA and for preserving limited samples of precious stock material. Next-Generation Sequencing (NGS) techniques can benefit from WGA, but due to their high sensitivity, WGA reliability needs to be certified to ensure an unbiased and accurate amplification of whole genomes. Myelodysplastic Syndromes (MDS) are a group of clonal hematopoietic stem cell disorders characterized by presenting somatic mutations in several myeloid-related genes. We have performed whole exome sequencing (WES) and targeted deep sequencing in tumoral samples from MDS patients. With the aim to determine if Multiple Displacement Amplification-based WGA can be applied to perform NGS in these type of samples and to obtain valuable results, targeted deep sequencing was performed on both fresh-DNA and WGA-DNA from the same patients.
Mehtods: Whole bone marrow samples from four MDS patients were included in the study. WGA was performed in tumoral DNA samples with REPLI-g (Qiagen). WES libraries were generated in tumoral-control paired samples using the SureSelect Human Exome Kit 51Mb v4 (Agilent) and sequenced on an Illumina HiSeq2000. Targeted sequencing libraries were prepared for fresh-DNA and WGA-DNA following the manufacturer specifications for TruSight Myeloid Sequencing Panel protocol (Illumina), and then sequenced on one single run on an Illumina MiSeq. WES sequencing data was analyzed using an in-house pipeline, as previously reported. Targeted sequencing data analysis was performed with theMiSeq Reporter Software (Illumina). Filtering was performed in all cases by eliminating sequencing and mapping errors and by discarding intronic or synonym variants, variants located at highly variable regions or with low coverage, as well as know polymorphisms. Additional filtering was performed by visualization on Integrative Genome Viewer Software v.2.3.72.
Results: Regarding targeted sequencing, fresh-DNA samples generated 6 million reads (SD = 1.9 million), with 98.5 % (SD = 0.8) of the mapped reads on-target and a mean target coverage of 12148.8 (SD = 3872.9). WGA-DNA samples yielded about 5.2 million reads (SD = 1.5 million), with 98.3 % (SD = 0.4) of the mapped reads on-target and a mean target coverage of 10447.5x (SD = 2946.3). A mean of 77% of total bases displayed a Q score ≥30, which did not differ between fresh and WGA-DNA.
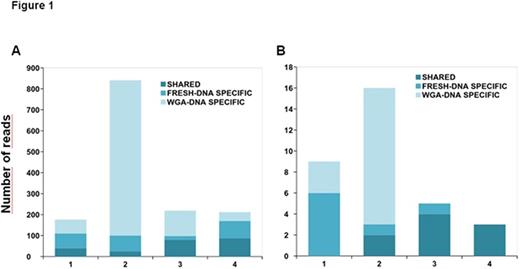
Comparison of all filtered variants within the four pairs revealed a high level of discordance between fresh/WGA samples (Figure 1A). A mean of 86% of the detected variants, considering both fresh and WGA-DNA, were detected at a low frequency (<10%). Therefore, a stricter variant filtering was performed, in which all variants detected at a frequency <10% were removed from further analyses.
The pairwise comparison across the paired samples showed a total of 48.1% (SD = 49.3) of common variants, 23.2% (SD = 30.1) of variants exclusively detected in fresh-DNA, and 28.7% (SD = 38.4) of variants exclusively detected in WGA-DNA (Figure 1B). Overall, 100% (n=9) of the common variants were also detected by WES. Regarding fresh-DNA specific variants, 63% (5/8) were seen by WES and 37% (3/8) were not. However, these three variants were detected by targeted sequencing at frequencies between 10-12%. This suggests that even a stricter filtering may be necessary when working with WGA-DNA, or that they were not detected by WES because it was performed at a mean coverage of 60x making it difficult to detect low frequency variants. None of the WGA-DNA specific variants were seen by WES. Taking all these factors into account, we used the fresh-DNA specific variants as the gold standard to further calculate the Positive Predictive Value (PPV) and the sensitivity of the WGA-DNA samples, and thus validate the accuracy of WGA technique in the sample preparation. This revealed a sensitivity of 61.7% (SD = 43.3) and a PPV of 53.3% (SD = 54.2).
Conclusions: These findings suggest that WGA methods may introduce errors, that can be detected at a low frequency, and that some bias can be expected, explaining why some variants present the gDNA may be lost during the amplification process. Therefore, we believe that applying WGA before library preparation should be restrained to cases with very limited material source and should be followed by a more in-depth and strict bioinformatics analysis and filtering process.
Sole:Celgene: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal