Abstract
Background
Aberrant patterns of alternative splicing (AS) are reported in acute myeloid leukemia and other malignances, but have not yet been investigated in myelodysplastic syndrome (MDS). In addition, mutations in spliceosome genes are especially common in MDS. Therefore, it would be highly interesting to analyze the clinical and biological significance of aberrant AS in MDS.
Materials and Methods
A total of 243 newly diagnosed MDS patients, including 78 with refractory anemia (RA), 22 with RA with ring sideroblasts(RARS), 76 with RA with excess blasts (RAEB), 24 with RAEB in transformation (RAEBT), and 43 with chronic myelomonocyticleukemia (CMMoL), who had complete clinical and genetic information were enrolled. We also included 20 healthy donors of hematopoietic stem cell transplantation for comparison. We extracted RNA from mononuclear cells (MNC) isolated from the bone marrow (BM), followed by hybridization on the microarrays of AffymetrixHuman Transcriptome Array 2.0, which contained > 6 million distinct probes covering coding, non-coding transcripts, and the exon-exon junctions among 67539 genes (44710 coding genesand 22829 noncoding genes). This comprehensive coverage of exons and junctions facilitated investigation of the splicing patterns of the genes. The aberrant AS was analyzed with the splicing index (SI) method using the default setting of AffymetrixTranscriptome Analysis Console version 3.0 software. The SI of a probe in a gene was derived from the ratio between MDS patients and normal controls in terms of the probe intensity divided by the gene expression level, ie. [intensityof probe A in gene A /gene A expression level]MDS/[intensity of probe A in gene A /gene A expression level]normal control. SI had to be less than -2 or more than 2 to be defined as an aberrant AS event.
Results
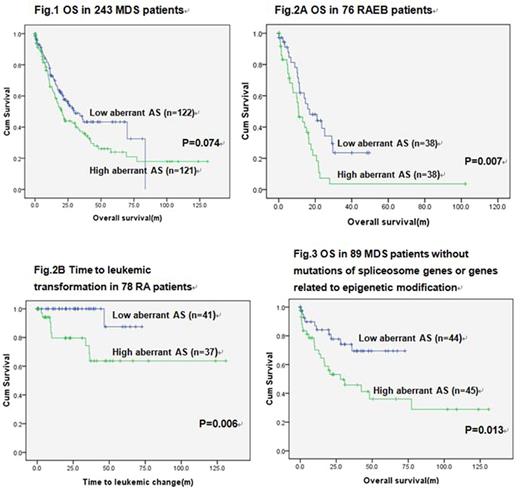
Totally 52730 of 67539 genes (78.1%) on the array were expressed in BM from both MDS patients and normal donor controls. Approximately 17565 of the 52730 expressed genes (33.3%) were differentially spliced between the MDS and normal marrow MNC (P < 0.05). As a whole, 88.9% of these aberrant spliced events were mapped to coding genome and 11.1% to noncoding regions. These aberrant AS exons fell into five common patterns, including exon skipping (57.8%), alternative donor sites (18.1%), alternative acceptor sites (17.8%), intron retention (5.6%) and mutually exclusive exons (0.7%). In average, when compared with the normal marrow MNC, there were 6.4 (4.8-12.0) more aberrant AS events in each AS gene (ASG) in the marrow MNC of 243 MDS patients.More aberrant AS events per ASG implied a more complex aberrant AS pattern.The numbers of aberrant AS events per ASG were not significantly associated with MDS subtypes and frequencies of spliceosome gene mutations. However, with a median follow up of 48.4 months, the patients with more aberrant AS events per ASG had a trend of shorter overall survival (OS) (median 20.1 vs 29.6 months, P=0.074) and a trend of shorter time to acute leukemic transformation (P=0.212) than the others (Fig.1). Further subgroup analysis showed a significantly shorter OS in the patients with more aberrant AS events per ASG than those with less events among RAEB patients (median OS: 10.8 months vs 16.9 months, P=0.007, Fig.2A). In addition, RA patients with more aberrant AS events per ASG had shorter time to acute leukemic transformation than RA patients with fewer aberrant AS events per ASG (P=0.006, Fig.2B). Among the patients without detectable mutations in spliceosome genes or genes related to epigenetic modification, those with more aberrant AS events per ASG also had a shorter OS than the others (median OS: 27.9 months vs not reached, P=0.013, Fig.3). Multivariate analysis in our cohort of 243 MDS patients revealed that higher aberrant AS events per ASG was an unfavorable prognostic factor for OS (P=0.016) independent of age, IPSS-R risk score, and mutations of SF3B1, ASXL1 and TP53.
Conclusion
To our knowledge, we are the first to present the prognostic impact of aberrant AS pattern in MDS patients. Large prospective cohorts are needed to confirm our observations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal